Abstract
In machine learning, hyperparameter tuning is strongly useful to improve model performance. In our research, we concentrate our attention on classifying imbalanced data by cost-sensitive support vector machines. We propose a multi-objective approach that optimizes model’s hyper-parameters. The approach is devised for imbalanced data. Three SVM model’s performance measures are optimized. We present the algorithm in a basic version based on genetic algorithms, and as an improved version based on genetic algorithms combined with decision trees. We tested the basic and the improved approach on benchmark datasets either as serial and parallel version. The improved version strongly reduces the computational time needed for finding optimized hyper-parameters. The results empirically show that suitable evaluation measures should be used in assessing the classification performance of classification models with imbalanced data.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Classification problems may be encountered in different domains. One of these is the disease diagnosis, which establishes the presence or absence of a given disease according to referred symptoms and results of medical exams. Machine learning approaches can be employed to support experts in diseases diagnosis. Many researches aim to propose new methods to improve or enhance the outcomes of existing ones.
Support vector machines (SVM) are one of the best machine learning (ML) models for solving several real-life classification problems (Vapnik 1998; Cristianini and Shawe-Taylor 2000). The choice of hyper-parameters of a ML model can significantly affect the resulting model’s performance. Generally, hyper-parameters are adjusted for each model in order to find a hyper-parameter setting that maximizes the model performances and so that the ML model can predict unknown data accurately. The goal of hyper-parameter optimization is to find a set of values that minimizes a predefined loss function.
Usually, a good set of hyper-parameters are determined by a grid search. The grid search strategy is based on testing all hyper-parameter combinations specified in a multi-dimensional grid. During the search, the hyper-parameters are varied, with fixed step-size, in a given range of values. The performance of a combination of hyperparameters is evaluated using a performance metric. The configuration with the best performance is selected and used to train the ML model on the whole dataset. However, this kind of search is very time consuming and it is suitable for the adjustment of few hyper-parameters.
Another big challenge in data mining that is attracting increasing interest of researchers is dealing with imbalanced data sets (Japkowicz and Stephen 2002). A dataset is imbalanced when one or more classes have very low proportions in the data as compared to the other classes. The first class is called as minority class with respect to the majority class(ess). The main interest is in correctly classifying the minority class. The existing methods for classification of imbalanced data can be categorized as algorithm-level category, data-level category, and cost-sensitive methods that lie between the above two categories (Galar et al. 2012). The first category includes methods modified or designed to handle imbalanced data; the second category includes methods that try to transform data in order to balance classes and use then standard classification algorithms. Down-sampling approaches, which reduce the majority class in the training subset, and over-sampling approaches, which increase the size of the minority class in the training subset, belong to this category. Finally, the third category includes methods designed for weighting differently the classes by introducing misclassification costs.
It is important to point out that the most commonly used model evaluation metric is the accuracy. However, it can be very misleading when data are imbalanced. In such cases, different evaluation metrics should be considered. We tested in (Guido et al. 2021) two evaluation model metrics, i.e., accuracy and G-Mean, on two imbalanced benchmark datasets by optimizing hyper-parameters of support vector machines by genetic algorithms (GAs). Comparing the results, we observed empirically that G-Mean is more suitable than accuracy to evaluate model performance in case of imbalanced data, especially when data refers to medical domains, like diagnosis. The results encouraged us to continue exploring this research field.
This research paper addresses the optimal hyper-parameters problem as a multi-objective problem. It has a twofold contribution:
-
1.
The main goal is to investigate methods for improving hyper-parameter tuning of SVM. We propose a novel approach for optimal hyper-parameter tuning that consists of a genetic algorithm combined with a decision tree. The basic idea is that some chromosomes are similar among them and they have thus the same fitness value. A decision tree (DT), trained in a suitable manner, is exploited to reduce the number of k-fold cross-validation to be performed and thus the overall computational time. As we will see, GAs were chosen even because they allow for an easy parallelization of the problem, which is tremendously helpful. The approach that combines GAs and DT strongly reduces the overall computational time, as described in Sect. 5.
-
2.
It focuses on testing and optimizing, at the same time, more suitable performance measures in addition to the accuracy. This is important for application domains where one data class is of more interest than others.
The paper is structured as follows. A short review of the state-of-the-art of the literature focusing on imbalanced data sets is in Sect. 2. We give a short description of support vector machines and decision trees in Sect. 3, and discuss some metrics commonly used to evaluate model performance. In Sect. 4, we introduce multi-objective optimization problems and the Non-dominated Sorting Genetic Algorithm-II (NSGA-II). In Sect. 5, we detail our approach that combines genetic algorithms and a heuristic procedure based on decision tree in order to find optimal hyper-parameters. Three objective functions are optimized. We perform several computational experiments aimed at finding the best hyper-parameter tuning for six benchmark datasets. The best results along with a discussion and comparison with other results of the literature are reported in Sect. 6. Finally, the conclusions are given in Sect. 7.
2 Related work on imbalanced data classification and cost-sensitive learning problems
Let \(D=\{(x_1, y_1), (x_2, y_2), \ldots , (x_l, y_l)\}\), be a dataset where \(x_i \in \Re ^L\) is a pattern (even called example) drawn from a domain X and \(y_i \in Y\) is its related class label. An example is thus a vector. In a binary classification domain, an example can be either positive, denoted by a label \(y=1\), or negative, denoted by \(y=-1\). Generally, the goal of a binary classifier is to map feature vectors \(x \in X\) to class labels \(y \in \{\pm 1\}\). In terms of functions, a classifier can be written as \(h(x) = sign[p(x)]\), where the function \(p : X \rightarrow R\) is denoted as the classifier predictor.
Classifiers generally perform poorly on imbalanced datasets and, as a consequence, often they classify almost all instances as negative. In recent years, imbalanced data classification has been studied by many researchers with different methods (Jo and Japkowicz 2004; Galar et al. 2012). These methods can be distinguished into two categories based on data and algorithms. Data-based methods focus on data pre-processing to reduce imbalanced data. For instance, up-sampling and under-sampling are two methods that modify instance distribution. Up-sampling methods increase the minority samples, whereas under-sampling methods reduce the majority samples. Synthetic Minority Oversampling Technique is an oversampling method that balances data by generating new samples similar to the minority samples and their neighbors (Chawla et al. 2002).
Hereafter, a positive instance belongs to the minority class, whereas a negative instance to the majority class. In many real-world applications, misclassifications may have different costs, such as for instance disease diagnosis and business decision making. The related classification problem, called cost-sensitive learning problem, aims at minimizing the total misclassification costs. The issue of classifying imbalanced data by an SVM was addressed in (Veropoulos et al. 1999) by a biased-SVM. This method uses two penalty coefficients for misclassified positive instances and negative instances. Since the positive instances usually belong to the minority class, the used penalty coefficient for this class is bigger than the penalty coefficient associated with the majority class. In this way, the SVM classifier aims at reducing misclassification rate of the minority class.
The performance of an SVM model even depends, for instance, on the used kernel function, which maps instances-vectors from the original input space to higher dimensional spaces to deal with nonlinearly separable data (Scholkopf and Smola 2001). Accordingly, two parameters of SVM, i.e., C and the kernel parameter were found by an exhaustive search approach in (Mehrbakhsh et al. 2019). Iranmehr et al. (2019) extended the SVM with cost-sensitive learning considering example dependent costs. They performed experimental analysis on class imbalance, cost-sensitive learning with a given class and example costs and showed that their proposed algorithm provides superior generalization performance compared to conventional methods. Qi et al. (2013) proposed a new Cost-Sensitive Laplacian SVM and tested its effectiveness via experiments on public datasets. They evaluate the algorithms performance by the Average Cost. Tao et al. (2019) developed a novel self-adaptive cost weights-based SVM cost-sensitive ensemble for imbalanced datasets classification tasks. The approach was tested on synthetic datasets and on public datasets showing higher classification accuracy than the other existing imbalanced classification methods in terms of G-Mean and F-Measure.
Evolutionary algorithms are flexible and commonly used for a plethora of machine learning problems and tasks (Bergstra et al. 2011; Goldberg and Holland 1988). Evolutionary optimization-based techniques solve the filter design task as an optimization problem. They are used successfully in different real-world optimization problems related to Finite Impulse Response (FIR) and Infinite Impulse Response (IIR) digital filters design. The goal is to minimize an error function that quantifies deviation between a filter and a desired response. This error is reduced by updating iteratively a set of filter coefficients such that given specifications are met. Dwivedi et al. (2018) provided a comprehensive review of the various evolutionary optimization-based techniques for FIR filter design. Approaches to design IIR filters based on evolutionary techniques were proposed in (Agrawal et al. 2018, 2017). Evolutionary algorithms are even used to automatically tune several parameters. Lessmann et al. (2005) used a GA in order to tune SVMs. Phienthrakul and Kijsirikul (2010) improved the accuracy of SVM by a nonlinear combination of multiple RBF kernels to obtain more flexible kernel functions. The hyperparameters are chosen by an evolutionary strategy where the objective functions are based on training accuracy, bounding of generalization error, and subset cross-validation on training accuracy. The resulting kernel allows better discrimination in the feature space than that of a single RBF kernel.
One of the first research papers on cost-sensitive approach tackled with an evolutionary process is due to Turney (1995). Recently, Noia et al. (2020) applied SVM, k-Nearest Neighbors and k-means as clustering techniques to predict the probability of contracting a given disease starting from both workplace-related (using Ateco and Istat codes) and worker-related characteristics (i.e., age at hiring, age at disease certification, gender, employment duration). They used a GA to find the best values of the used methods. Misclassification error rate is used as fitness function. However, since the classes were not evenly distributed among the instances, they used a second fitness function that reduces the misclassification error rate of the minority class.
An exhaustive search of papers addressing evaluation of ML algorithms on classification is due to Sokolova et al. (2006). These authors showed that the clear “leaders” are those papers in which evaluation is performed on data from the UCI repository, in biomedical and medical sciences, visual and text classification, and language applications. The most used evaluation measures are accuracy, precision, recall, F-score, and the Receiver Operating Characteristic (ROC).
3 Learning model classifiers
We optimize hyper-parameters of SVM classifiers with Gaussian kernel in order to correctly compare our results found on public and well-known datasets with those reported in the literature. Our approach, as it is better detailed in Sect. 5, trains and uses random trees to reduce the overall computational time.
In this section, we briefly introduce SVM and decision trees. Thus, we report the most used performance metrics of a ML model and discuss their suitability in case of imbalanced datasets.
3.1 Support vector machine
The SVM was introduced by Cortes and Vapnik (1995) and is based on statistical learning theory (Vapnik 1998). SVMs are a class of algorithms for classification, regression and other applications (Cristianini and Shawe-Taylor 2000) and they are among the most used ML techniques.
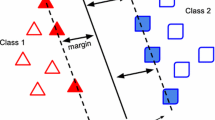
Let X, be a dataset with L instances \(X=(x_1, \ldots , x_L)\), where \(x_i \in \Re ^m,\) denotes an instance with m features, and \(y_i \in \{\pm 1\}\) its label, \(i=1,\ldots , L\). In a binary classification problem, an SVM basically searches for an optimal hyperplane that separates patterns of the two classes by maximizing the margin \(w \in \Re ^m\). Finding the optimal hyperplane means solving the quadratic programming model (1)-(3), which is known as soft-margin SVM
where C, named penalty parameter, is a trade-off between the size of the margin of separation w and the training errors \(\xi \); b is the bias and it indicates the offset of the hyperplane from the origin. Constraints (2) state that when a training example \(x_i\) lies on the wrong side of the hyperplane, the corresponding slack variable \(\xi _i\) is greater than 1. Small values of C increase the training errors, whereas larger values bring it closer to the hard-margin SVM. In case of nonlinearly separable datasets, the SVM basically maps input vectors into high-dimensional feature spaces by the so-called kernel functions (Hofmann et al. 2008). A kernel function, denoted as \(K(x_i,x_j) = \bigl \langle \phi (x_i),\phi (x_j)\bigr \rangle \), is an inner product in a feature space where it measures similarity between any pair of inputs \(x_i\) and \(x_j\). A kernel function can take many different forms (Hofmann et al. 2008), such as
-
Linear kernel \(K(x_i, x_j)=(x_i^T x_j)^d\)
-
Polynomial kernel \(K(x_i, x_j)=(x_i^T x_j + a)^d\)
-
Radial Basis Function (RBF) kernel \(K(x_i, x_j)=exp(-\gamma \Vert x_i - x_j\Vert ^2)\)
The decision function, i.e., the classifier, is specified by a subset of training instances, the so-called support vectors, that are the only vectors that “support” the optimal separating hyperplane.
It is well known that the performance of most machine learning algorithms on a given dataset depends on well-tuned hyper-parameter. In setting up an SVM model, for instance, two problems are encountered: (1) how to select the kernel function, and (2) how to select its hyper-parameter. An SVM with polynomial kernel has three parameters that need to be optimized: the regularization parameter C, the parameter a, and the degree d. The optimization of these three parameters if 50 steps should be performed, requires an amount of time to test the total \(50^3= 125000\) combinations. The greater the number of parameters to be set, the greater is the number of combinations.
The cost-sensitive SVM (CS-SVM) uses two penalty weights for the two classes. Let \(C_1\), be the cost of a false negative. It penalizes misclassification of instances of the minority class. Analogously, let \(C_{-1}\), be the cost of a false positive. It penalizes misclassification of instances of the majority class. The optimization model CS-SVM is (4)-(6).
Observe that the cost matrices has the diagonal elements as zero— because of the assumption that a correct classification has no cost—and the off-diagonal elements are positive numbers. However, the range of possibilities for CS-SVM hyper-parameter can be huge.
Datta and Das (2015) proposes a Near-Bayesian Support Vector Machine (NBSVM) for imbalanced classification problems by combining decision boundary shift and unequal regularization costs. Extensive comparison with standard SVM and some state-of-the-art methods is furnished as a proof of the ability of the NBSVM to perform competitively on imbalanced datasets.
3.2 Decision tree
A decision tree is a supervised learning algorithm for regression and classification problems (Breiman et al. 1984) and is the most popular form of rule-based classifiers (Witten and Frank 2005). It has a set of elements called nodes and is built top-down from a root node. Each node represents a single input attribute: leaf nodes contain an output attribute, which is used to make a prediction; the other nodes are split points of an attribute. The data is partitioned into homogeneous subsets, i.e., they contain instances with similar values. Given a new input, the tree is traversed by evaluating the specific input started at the root node of the tree.
3.3 Performance evaluation and some limitations
To estimate the generalization performance of an SVM model, generally one evaluates accuracy measure on data not used for training the model. The k-fold cross-validation (k-CV) is the most used procedure. It consists on partitioning data into k disjoint sets of approximately equal size. An SVM is thus trained k times: at the \(i-th\) iteration, all the disjoint sets are used as training set except the \(i-th\) set, which is used to evaluate the performance of the model. The errors observed in this process are averaged yielding the k-fold CV error.
Before introducing the most used evaluation measures, it is useful to revise the confusion matrix of binary classification problems. A general confusion matrix is illustrated in Table 1. The two columns refer to the predicted classes, whereas the two rows refer to the actual classes. True Positives (TP) is the number of positive instances correctly classified and False Negatives (FN) is the number of positive instances incorrectly classified as negative. These two numbers refer to the minority class. Similarly, True Negatives (TN) is the number of negative instances correctly classified, and False Positives (FP) is the number of negative instances incorrectly classified as positive class. These two numbers refer to the majority class. Observe that, in case of data related to patients, a false negative means that patient has the disease but the diagnosis result says that it does not have.
The most common evaluation measures used are listed below.
Accuracy defined as the ratio between the number of instances correctly classified and the total number of instances. It assesses the overall effectiveness of the model by showing the probability of the true value of the class label
Other two measures that separately estimate a classifier’s performance on different classes are sensitivity and specificity. They are often employed in medical and bio-medical applications.
Sensitivity (true positive rate) is defined as the ratio between the number of positive instances correctly classified as such and the number of positive instances
Specificity (true negative rate) is defined as the ratio between the number of negative instances correctly classified as such and the number of negative instances
Precision is defined as the ratio of TP to the number of all instances predicted as positive
As reported especially recently in some papers (e.g., Tao et al. 2019), the accuracy-based evaluation measure is not suitable for classification of imbalanced data as the minority class has very little effect on the accuracy compared to the majority class. For imbalanced classification problems, the correct classification of instances of the minority class is usually the most important measure. There are further interesting classification evaluation measures that allow to balance false positive rate and false negative rate. Here, among these measures, we evaluate even F-Measure, the Geometric Mean, the average cost, the Youden’s index, and the balanced accuracy. They are defined as follows.
F-Measure integrates sensitivity and precision into an average by a harmonic mean
The harmonic mean of two numbers tends to be closer to the smaller number. A high F-Measure value means that both Sensitivity and Precision are high.
Geometric Mean(G-Mean) is suggested as the balanced performance between the two classes. It is intrinsically defined as the geometric mean of sensitivity and specificity. If the G-Mean value is high, both Sensitivity and Specificity are expected to be high simultaneously
Average Cost(AC) is expressed as
where \(C_1\) and \(C_{-1}\) are the two costs used in the objective function of CS-SVM.
Youden’s index Y equally weights the algorithm’s performance on positive and negative instances:
Balanced accuracy(BA) is the average of sensitivity and specificity:
4 Multi-objective optimization problems and Genetic algorithms
Multi-objective optimization problems consist of more than one criterion, often conflicting, for which any solution existing on the Pareto front of criterion trade-offs is considered optimal.
In this section, we introduce multi-objective optimization problems and the cornerstone concept of Pareto optimality.
A multi-objective problem consists of minimizing and/or maximizing two or more objective functions subject to inequality and/or equality constraints. The objective functions are conflicting among them and a solution is a trade-off in the objective function space.
Definition 1
A solution is defined Pareto optimal if there does not exist any other solution in the objective space which improves the value of any of its objective functions without deteriorating at least one other objective function value.
In other words, a non-dominated solution provides a suitable compromise between all objectives without degrading any of them. The multi-objective optimization process is looking for a set of alternative solutions that represent the Pareto optimal solution. A set of non-dominated individuals form a Pareto-optimal front.
From the mathematical point of view, the definition of the dominance between two solutions \(x_1\) and \(x_2\) is that \(x_1\) is no worse than \(x_2\) in all objectives \(f_i, i \in \{1, \ldots , m\}\) of the problem. This concept can be expressed as \(x_1\) dominates \(x_2\) if \(f_i(x_1) \le f_i(x_2) \ \forall i \in \{1, \ldots , m\} \ and \ \exists \ j \in \{1, \ldots , m\}:f_j(x_1) \le f_j(x_2)\).
The genetic algorithms were developed by Holland and his collaborators (Holland 1975) as a model based on Charles Darwin’s theory of natural selection. They are heuristic search techniques, successfully applied to different domains (e.g., Guido and Conforti 2017; Bao-De et al. 2021). Furthermore, they demonstrated a large amount of inherent parallelism that makes them attractive mainly for solving problems defined in large feature spaces, as that one here addressed. The evolutionary process usually starts from a population of randomly generated individuals, which are the chromosomes. It is an iterative process. One iteration is one generation. In each generation, the fitness of every individual in the population is evaluated. The fitness value of a chromosome is a measure of its goodness. The fitness is usually the value of the objective function in the optimization problem being solved. Usually, operators such as selection, crossover, mutation and recombination are applied during the evolutionary process over the generated populations to find better chromosomes, which optimize the fitness function till a termination condition is reached. The offsprings in a population act like independent agents so that they explore the search space in many directions.
As well known, genetic algorithms have some disadvantages mainly due to the choice of parameters such as the mutation rate and crossover rate that should be carried out carefully. The crossover operator is one of the most important operators because it determines the global convergence of the genetic algorithm.
4.1 NSGA-II
Srinivas and Deb (1994) proposed an algorithm based on non-dominated sorting for solving multiobjective problems. This algorithm was called non-dominated sorting genetic-algorithm (NSGA). Deb et al. (2002) improved it by proposing NSGA-II. The key features of NSGA-II are elitism, diversity-preserving mechanisms, and emphasis on non-dominated solutions. In NSGA-II, the N offsprings are created from the N parents using standard genetic algorithms. The new population at the next generation is given by selecting the non-dominated solutions for the Pareto front with the highest diversity while discarding the rest of the solutions.
Tournament selection This is a procedure that imitates survival of the fittest in nature. Indeed, each individual competes in two tournaments with randomly selected individuals. The crowded tournament selection is based on ranking and distance: if a solution has a better rank than another one, it will be selected; if the ranks are the same but the crowding distance is not, the solution with better crowding distance is selected.
Crowding distance The crowding distance metric of an individual proposed by Deb and Goel (2001) aims to select potential individuals to construct a new population. It is essentially based on the cardinality of a solution sets and their distance to solution boundaries. More specifically, it is defined as the perimeter of the rectangle with its nearest neighbors at diagonally opposite corners. Two individuals with a same rank are better if they have a larger crowding distance.
Crossover and mutation Crossover and mutation are employed to obtain the offspring population.
Algorithm 1 shows the framework of NSGA-II. The main steps of NSGA-II can be summarized as follows:
-
Step 1
Create a new population by combining parents and offsprings and apply non-dominated sorting
-
Step 2
Identify different fronts
-
Step 3
Generate the new population by exploiting the fronts given at the previous step until size N
-
Step 4
Use the crowd distance to carry out a crowding sort applied to the fronts
-
Step 5
Generate new offspring from the current population via the genetic operators crossover, mutation, and selection

5 Proposed approach
In this section, is firstly introduced a basic approach for hyper-parameter optimization. Then, a novel algorithm for hyper-parameters tuning based on GA and DT is proposed. The core of the algorithm is a fitness function evaluation procedure along with a similarity procedure.
5.1 Basic approach
The basic approach consists on using NSGA-II algorithm for solving a multi-objective hyper-parameter tuning problem. A set of hyper-parameter codified as a chromosome is evaluated by a k-fold CV approach. A fitness function evaluation is thus performed at each generation, i.e., each chromosome has its fitness functions evaluated. However, this approach is quite time consuming. Indeed, let N, be the number of chromosomes of a population, and G the number of generations. At each generation, the number of carried out k-fold CV is N, one per each chromosome. The overall number of performed k-fold CV is thus \(N \times G\). For example, if \(N=24\) and \(G=200\), the overall number of k-fold CV to be carried out is 4800 and the computational time may be extremely high.
There are two main issues: the first one is related to the time needed to carry out k-fold CV; the second one, is related to the fact that often a chromosome is slightly different from another one already evaluated and with equal fitness. We try to overcome these two issues by introducing a procedure in the NSGA-II algorithm that exploits a suitable and trained DT. The proposed algorithm, described in the following, reduces considerably the overall number of performed k-fold CV by combining NSGA-II with a DT. The goal is to evaluate only a small set of chromosomes at each generation by a k-fold CV. This procedure does not affect convergence of the algorithm and strongly reduces the overall computational time.
5.2 Improved hyper-parameters algorithm
The above basic approach has been modified in order to evaluate the fitness function only of some individuals of a population by a k-fold CV. Figure 2 provides an intuitive understanding of the proposed algorithm framework.
Each chromosome consists of a number of genes that represent the hyper-parameters of CS-SVM. The algorithm starts from an initial population \(Pop_0\). It consists of the following five main steps.

The core of Algorithm 2 is the fitness evaluation procedure at Step 2, explained in the following.
Step2: Fitness evaluation procedure The aim of the fitness evaluation step is to provide a procedure that reduces the number of fitness evaluations and consequently the number of carried out k-fold CV. To this purpose, a DT is trained at each generation and used to predict the fitness value of some chromosomes, as explained below. Indeed, the fitness of a chromosome in a population is evaluated or assigned: A whole population is evaluated by k-fold CV only at those generations well-defined in the set GenSet. This means that the cost-sensitive learning classifier SVM-based is built using the hyper-parameters codified as chromosomes of the population; for every chromosomes, a k-fold CV is used to estimate the generalization ability of the related build model. The set GenSet has at least two elements, i.e., the first and the last generation. A procedure based on a learned DT takes place at those generations not in the set GenSet.
The fitness evaluation procedure is depicted in Fig. 3. To reduce the overall computational time, the procedure verifies if each chromosome has already a fitness value (because it has been evaluated previously). If so, the procedure analyzes next chromosome; otherwise, the chromosome is compared, at Step 2.2, with the chromosomes of the previous population in order to discover similarity. If the chromosome is similar at least to one chromosome, the DT trained on the previous population predicts its fitness value; this value is thus assigned as predicted value. Otherwise, if no similarity is found, a cost-sensitive learning classifier SVM-based is built and the fitness value is evaluated by k-fold CV.
Similarity between two chromosomes can be estimated by various distance measurement methods. Here, we designed a procedure that evaluates similarity between two chromosomes as follows. Let \(chr_1\) and \(chr_2\), be two chromosomes represented as vectors. The procedure compares each corresponding couple of genes of \(chr_1\) and \(chr_2\), as detailed in Algorithm 3. More specifically, the difference between the \(i-th\) gene of \(chr_1\) and the corresponding gene of \(chr_2\) is computed. If this difference is less than a given threshold \(t_i\), the next couple of genes of the two chromosomes are compared; otherwise, the procedure stops and the two chromosomes are not similar.
Figure 4 depicts an example of DT trained to predict a given fitness function.

6 Experimental results and analysis
In this study, we test the proposed Algorithm 2 for on six benchmark imbalanced datasets binary classification task to compare the performance of different classification methods in the literature with our results. They are related to medical diagnosis represented as binary classification problems and have different sample sizes, attributes, and imbalance ratio (IR), defined as m/M (Amin et al. 2016), where m is the number of the minority instances and M is the number of majority instances.
We conducted experiments to answer the following research questions empirically:
-
1.
Does multi-objective optimization find much sparser solutions without a major loss in predictive performance compared to single-objective optimization?
-
2.
Are there alternative metrics to the accuracy?
-
3.
May the computational time be reduced by a machine learning technique?
A brief description of the datasets is in Sect. 6.1. Details on the algorithms embedded in our approach and the hyper-parameter spaces of the several CS-SVM that are being investigated and tuned over are reported in Sect. 6.2. Experimental results are listed in Sect. 6.3.
6.1 Benchmark datasets
The datasets are from the University of California Irvine (UCI) Repository of Machine Learning Databases (https://archive.ics.uci.edu/ml/datasets.php). They have diversity in the number of attributes and imbalance ratio. Moreover, the datasets have both continuous and categorical attributes, and some of them have missing values.
Appendicitis dataset consists of 106 instances and 8 attributes. The attributes are results of laboratory test.
Haberman dataset describes the five-year or greater survival of breast cancer patients. The study was conducted between 1958 and 1970 at the University of Chicago’s Billings Hospital. The dataset consists of 306 instances and 4 attributes. The outcome is patient survival. There are no missing values.
Hepatitis dataset is used to classify patients with hepatitis in the two classes, live or die. It consists of 155 instances and 19 attributes, 14 nominal attributes and 6 multi-valued attributes. It requires the determination of whether patients with hepatitis will either live or die. The problem aims to predict the presence or absence of hepatitis by using the results of various medical tests carried out on a patient. The dataset has missing values.
Pima Indian Diabetes dataset is used to predict whether or not a patient has diabetes. All patients are female, are at least 21 years old, and are of Pima Indian heritage. It has 8 laboratory features.
Wisconsin Diagnostic Breast Cancer (WDBC) dataset consists of 30 features computed by digitized image of fine needle aspirate of a breast mass index. The problem aims to predict whether or not the patient has breast cancer.
Wisconsin Prognostic Breast Cancer (WPBC) dataset has 198 instances that represent follow-up data for one breast cancer case, only those cases exhibiting invasive breast cancer and no evidence of distant metastases at the time of diagnosis. It is used in this paper to classify patients as recurrences before 24 months (positive class) or non-recurrence beyond 24 months (negative class). We removed the feature named “Time” from the dataset because it is the recurrence time for instances in the positive class and the disease-free time for the instances of the negative class.
Table 2 summarizes, per each dataset, the number of attributes, the number of minority instances (diseased examples), the number of the majority instances (non-diseased examples), and the index ratio.
6.2 Learning algorithms and hyperparameters optimization
We considered several model classifiers CS-SVM with Gaussian kernel tuned by the optimization algorithm proposed. The experiments were performed by the ML algorithms of Waikato Environment for Knowledge Analysis (WEKA). WEKA is an open-source collection of ML algorithms and data processing tools. We used Sequential minimal optimization algorithm for SVM and Random Tree algorithm for DT. For that concerning NSGA-II algorithm, we used the framework named Java Class Library for Evolutionary Computation (JCLEC) Ramírez et al. (2015, 2019), which is a Java suite for solving multi-objective optimization problems using evolutionary algorithms.
Algorithm 2 has been coded in Java using the NSGA II algorithm of the JCLEC framework. We executed both the sequential and the parallel version of the NSGA-II. The parallel version is more efficient since it performs function evaluations of different individuals in parallel. The experiments were run on a PC Intel Xeon E5 1620 CPUs with 4 cores at 3.50 GHz and 32 GB RAM.
6.2.1 Parameter setting
Algorithm 2 starts from an initial population \(Pop_0\) of chromosomes randomly generated. Each chromosome has four genes representing the hyper-parameter of CS-SVM, as depicted in Fig. 5. All experiments have the same random initial population; the number of generations is the only one stop** criterion.
Table 3 lists the search population size, crossover probability \(p_c\), gene mutation probability \(p_m\), number of generations along with the design parameters (decision variables) and the range of their variations. We tested two population sizes and created the initial parent population randomly by selecting solutions from the ranges defined for the parameters \(C, C_{1}, C_{2}, \gamma \), where \(C_1\) and \( C_2\) are the costs of the minority class and majority class, respectively.
The multi-objective problem that we formulated and solved has three fitness functions (7–9), given by accuracy, G-mean, and Average Cost, respectively:
All the experiments are conducted by 10-fold cross-validation.
6.3 Computational results
To assess our approach, we performed both Algorithm 1 and Algorithm 2 on the six datasets and compared the results. The computational experiments were carried out using the JCLEC sequential algorithm and its parallelized version. The only difference we noticed was the reduced computational time of the parallelized version with respect to the sequential algorithm. We report in this section only the results found by the parallel version.
Table 4 reports the best fitness values per each dataset. From the second to the fourth column there is the value of accuracy, G-Mean, and average cost, respectively. We compare in this table our results with the best ones of the literature by selecting those papers that optimized hyper-parameter of SVM with RBF kernel. Under these conditions, experimental evidence shows that our algorithm finds similar results or outperforms the other algorithms proposed in the literature. The best results in terms of accuracy on Hepatitis, Pima, and WPBC datasets are found in (Yu and Wang 2017) by optimizing the parameters of the SVM with RBF kernel by a novel ensemble differential evolution approach that they proposed. Several approaches were tested in (Tao et al. 2019) and the results were reported in terms of G-Mean. In Table 4, we denoted with G-Mean\(^a\) and G-Mean\(^b\) the values found by CS-SVM and their self-adaptive cost weights-based support vector machine cost-sensitive ensemble approach, respectively. It is helpful to notice that they reported these results on the datasets by modifying imbalanced data ratio of 10:1.
Tables 5, 6, 7, 8 list only some of the found non-dominated solutions of the Pareto front of our experimental results. These results refer to the experiments carried out with the related parallelized version of Algorithms 1 and 2. The first and second column of these tables report the population size and the number of carried out generations; the next three columns show the fitness function values associated with the optimal hyper-parameter configuration, which is reported in the next four columns. The eleventh and twelfth column reports the sensitivity and specificity values, whereas the next four columns report the ROC area, the F-Measure, the balanced accuracy, and the Youden’s index, respectively. Finally, the last column shows the average computational time, in minutes.
As already observed in the literature, the accuracy is not a suitable measure for imbalanced data. Indeed, we noticed that in the Haberman dataset, for instance, there is a hyper-parameter configuration that allows to have a good accuracy equal to \(75.53\%\), but the specificity is zero as well as the G-mean value. Similar cases are on the Hepatitis and WPBC datasets.
Tables 5 and 6 show the results found by Algorithm 1 on the six datasets along with the related optimized hyper-parameter configuration. For the Appendicitis dataset, for instance, the best accuracy in Table 5 is \(f_1=89.62\); the best G-Mean is \(f_2=82.54\), and the best average cost is \(f_3=0.11\). As expected, the improvement of a fitness function implies a worsening in the other two. We observe that the single optimal fitness values are found with different hyper-parameter tuning. Moreover, the best results are found in all the experiments even if number of population size and generation number is increased.
Tables 7 and 8 show the results found by Algorithm 2 on the six datasets along with the related optimized hyper-parameter configuration. These results were found in very contracted computational time if compared to the previous ones. Observe that there has been a reduction over 70% in some experiments. These results show that the proposed Algorithm 2 is efficient.
The results evidenced that: (1) both algorithms converge and find the same best values for the three fitness functions; (2) the number of optimal non-dominated solutions of the Pareto front found by Algorithm 1 is greater than the number found by Algorithm 2. To better understand our finding, we illustrate in Figs. 6 and 7 the Pareto points of Tables 7 and 8 per each dataset with the six performance measures. The Pareto points are shown considering decreasing Sensitivity. As depicted in these two figures, generally balance accuracy decreases as Sensitivity decreases while Sensitivity increases. The best Pareto points related to medical datasets, as those tested in this paper, should be the points with high balance accuracy or high sensitivity values.
7 Conclusion
Support vector machines are one of the best ML models for solving several real-life classification problems. However, as in other ML techniques, their performance depends on hyper-parameters.
In this paper, we have investigated and proposed an approach that combines genetic algorithms and decision trees to optimize hyper-parameters of C-SVMs. The optimum values of the regularization parameter, costs of classes and the parameters of the RBF kernel function are searched for SVM.
We tested the algorithm on six benchmark datasets, which are imbalanced. We evaluated the performance of the models by several performance metrics. The framework is better or equivalent to other algorithms proposed in the literature for CS-SVM hyper-parameters optimization. Overall, taking into account three predictive metrics, i.e., accuracy, G-Mean, and average cost, the best hyper-parameter configuration is found in short computational time, mainly if compared with grid search approach. Hence, this approach can be considered as a good solution for addressing imbalanced dataset classification and hyper-parameter tuning, as they are challenging problems in classification research.
We suggest evaluating the performance of classifiers on medical data by suitable measures other than accuracy. Our future work is to extend and assess the proposed approach to investigate hyper-parameter tuning of different machine learning methods.
References
Agrawal N, Kumar A, Bajaj V (2017) A new design method for stable IIR filters with nearly linear-phase response based on fractional derivative and swarm intelligence. IEEE Transactions on Emerging Topics in Computational Intelligence 1(6):464–477
Agrawal N, Kumar A, Bajaj V (2018) Design of digital IIR filter with low quantization error using hybrid optimization technique. Soft Comput 22(9):2953–2971
Amin A, Anwar S, Aea Adnan (2016) Comparing oversampling techniques to handle the class imbalance problem: a customer churn prediction case study. IEEE Access 4:7940–7957
Bao-De L, **n-Yang Z, Mei Z et al (2021) Improved genetic algorithm-based research on optimization of least square support vector machines: an application of load forecasting. Soft Comput 10(1007):5674–9
Bergstra J, Bardenet R, Bengio Y, et al (2011) Algorithms for hyper-parameter optimization. In: and CAI (ed) Proceedings of the 24th international conference on neural information processing systems. USA, pp 2546–2554
Breiman L, Friedman JH, Olshen R, et al (1984) R. A. and Stone, C.J. Classification and regression trees. CRC press
Chawla N, Bowyer K, Lea Hall (2002) Smote: Synthetic minority over-sampling technique. J Artif Intell Res 16:321–357
Cortes C, Vapnik V (1995) Support-vector network. Mach Learn 20:273–297
Cristianini N, Shawe-Taylor J (2000) An Introduction to Support Vector Machines and other kernel-based learning methods. Cambridge University Press
Datta S, Das S (2015) Near-bayesian support vector machines for imbalanced data classification with equal or unequal misclassification costs. Neural Netw 70:39–52
Deb K, Goel T (2001) Controlled elitist non-dominated sorting genetic algorithms for better convergence. In: Lothar T, Kalyanmoy D, Coello C et al (eds) Zitzler Eckart. Evolutionary Multi-Criterion Optimization, Springer, Berlin Heidelberg, pp 67–81
Deb K, Pratap A, Agarwal S et al (2002) A fast and elitist multiobjective genetic algorithm: NSGA-II. IEEE Trans Evol Comput 6:182–197
Dwivedi AK, Ghosh S, Londhe ND (2018) Review and analysis of evolutionary optimization-based techniques for fir filter design. Circuits Syst Signal Process 37(10):4409–4430
Galar M, Fernandez A, Barrenechea E et al (2012) A review on ensembles for the class imbalance problem: Bagging, boosting, and hybrid-based approaches, systems, man, and cybernetics, part c: Applications and reviews. IEEE Trans 42(4):463–484
Goldberg DE, Holland J (1988) Genetic algorithms and machine learning. Mach Learn 3(2):95–99
Guido R, Conforti D (2017) Hybrid genetic approach for solving an integrated multi-objective operating room planning and scheduling problem. Comput Oper Res 87:270–282
Guido R, Groccia MC, Conforti D (2021) Hyper-Parameter Optimization in Support Vector Machine on unbalanced datasets using Genetic Algorithms. In: Optimization in Artificial Intelligence and Data Sciences, AIRO Springer Series (in press)
Hofmann T, Scholkopf B, Smola AJ (2008) Kernel methods in machine learning. Ann Statist pp 1171–1220
Holland JH (1975) Adaptation in natural and artificial systems: An introductory analysis with applications to biology, control, and artificial intelligence. Michigan Press
Iranmehr A, Masnadi-Shirazi H, Vasconcelos N (2019) Cost-sensitive support vector machines. Neurocomputing 343:50–64
Japkowicz N, Stephen S (2002) The class imbalance problem: a systematic study. Intell Data Anal 6:429–449
Jo T, Japkowicz N (2004) Class imbalances versus small disjuncts. ACM SIGKDD Explorations Newslett 6:40–49
Lessmann S, Stahlbock R, Crone R (2005) Optimizing hyperparameters of support vector machines by genetic algorithms. In: IC-AI pp 74–82
Mehrbakhsh N, Hossein A, Leila S et al (2019) A predictive method for hepatitis disease diagnosis using ensembles of neuro-fuzzy technique. J Infect Public Health 12(1):13–20
Noia A, Martino A, Montanari P et al (2020) Supervised machine learning techniques and genetic optimization for occupational diseases risk prediction. Soft Comput 24:4393–4406
Phienthrakul T, Kijsirikul B (2010) Evolutionary strategies for hyperparameters of support vector machines based on multi-scale radial basis function kernels. Soft Comput 14:681–699
Qi Z, Tiana Y, Shia Y et al (2013) Cost-sensitive support vector machine for semi-supervised learning. Procedia Comput Sci 18:1684–1689
Ramírez A, Romero JR, Ventura S (2015) An extensible JCLEC-based solution for the implementation of multi-objective evolutionary algorithms. In: proceedings of the companion publication of the 2015 annual conference on genetic and evolutionary computation, pp 1085–1092
Ramírez A, Romero JR, García-Martínez C et al (2019) JCLEC-MO: a java suite for solving many-objective optimization engineering problems. Eng Appl Artif Intell 81:14–28
Scholkopf B, Smola AJ (2001) Learning with Kernels: Support Vector Machines, Regularization, Optimization, and Beyond. MIT Press, Cambridge, MA, USA
Sokolova M, Japkowicz N, Szpakowicz S (2006) Beyond accuracy, F-score and ROC: A family of discriminant measures for performance evaluation. In: Sattar A, Kang B (eds) Advances in Artificial Intelligence. Lecture Notes in Computer Science, vol 4304. Springer, Berlin, Heidelberg
Srinivas N, Deb K (1994) Multiobjective optimization using nondominated sorting in genetic algorithms. Evol Comput 2(3):221–248
Tao X, Li Q, Guo W et al (2019) Self-adaptive cost weights-based support vector machine cost-sensitive ensemble for imbalanced data classification. Inf Sci 487:31–56
Turney PD (1995) Cost-sensitive classification: empirical evaluation of a hybrid genetic decision tree induction algorithm. J Artif Int Res 2:369–409
Vapnik V (1998) Statistical Learning Theory. Wiley, John Sons Inc
Veropoulos K, Campbell C, Cristianini N (1999) Controlling the sensitivity of support vector machines. In: proceedings of the international joint conference on AL, pp 55–60
Witten I, Frank E (2005) Data Mining Practical Machine Learning Tools and Techniques. Morgan Kaufmann Publishers, CA
Yu X, Wang X (2017) A novel hybrid classification framework using svm and differential evolution. Soft Comput 21:4029–4044
Acknowledgements
The research has been partially supported by the research project SI.F.I.PA.CRO.DE. Sviluppo e industrializzazione farmaci innovativi per terapia molecolare personalizzata PA.CRO.DE. (PON \(ARS01\_00568\), CUP: B29C20000360005, CONCESSIONE RNA-COR: 4646672), Italian Ministry of University and Research, 2021.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors of the manuscript declare that they have no affiliations with or involvement in any organization or entity with any financial interest (such as honoraria; educational grants; participation in speakers’ bureaus; membership, employment, consultancies, stock ownership, or other equity interest; and expert testimony or patent-licensing arrangements), or non-financial interest (such as personal or professional relationships, affiliations, knowledge or beliefs) in the subject matter or materials discussed in this manuscript.
Additional information
Communicated by Dario Pacciarelli.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Guido, R., Groccia, M.C. & Conforti, D. A hyper-parameter tuning approach for cost-sensitive support vector machine classifiers. Soft Comput 27, 12863–12881 (2023). https://doi.org/10.1007/s00500-022-06768-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00500-022-06768-8