Abstract
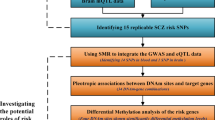
Recent genome-wide association studies (GWAS) have made substantial progress in identifying disease loci. The next logical step is to design functional experiments to identify disease mechanisms. This step, however, is often hampered by the large size of loci identified in GWAS that is caused by linkage disequilibrium between SNPs. In this study, we demonstrate how integrating methylome-wide association study (MWAS) results with GWAS findings can narrow down the location for a subset of the putative casual sites. We use the disease schizophrenia as an example. To handle “data analytic” variation, we first combined our MWAS results with two GWAS meta-analyses (N = 32,143 and 21,953), that had largely overlap** samples but different data analysis pipelines, separately. Permutation tests showed significant overlap** association signals between GWAS and MWAS findings. This significant overlap justified prioritizing loci based on the concordance principle. To further ensure that the methylation signal was not driven by chance, we successfully replicated the top three methylation findings near genes SDCCAG8, CREB1 and ATXN7 in an independent sample using targeted pyrosequencing. In contrast to the SNPs in the selected region, the methylation sites were largely uncorrelated explaining why the methylation signals implicated much smaller regions (median size 78 bp). The refined loci showed considerable enrichment of genomic elements of possible functional importance and suggested specific hypotheses about schizophrenia etiology. Several hypotheses involved possible variation in transcription factor-binding efficiencies.



Similar content being viewed by others
References
Aberg KA et al (2012) MBD-seq as a cost-effective approach for methylome-wide association studies: demonstration in 1500 case–control samples. Epigenomics 4:605–621. doi:10.2217/epi.12.59
Aberg KA et al (2013a) A comprehensive family-based replication study of schizophrenia genes. JAMA Psychiatry 70:573–581. doi:10.1001/jamapsychiatry.2013.288
Aberg KA et al (2013b) Testing two models describing how methylome-wide studies in blood are informative for psychiatric conditions. Epigenomics 5:367–377. doi:10.2217/epi.13.36
Aberg KA et al (2014) Methylome-wide association study of schizophrenia: identifying blood biomarker signatures of environmental insults. JAMA Psychiatry 71:255–264. doi:10.1001/jamapsychiatry.2013.3730
Adzhubei IA et al (2010) A method and server for predicting damaging missense mutations. Nat Methods 7:248–249. doi:10.1038/nmeth0410-248
Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M (2003) Proteomic characterization of the human centrosome by protein correlation profiling. Nature 426:570–574. doi:10.1038/nature02166
Benton CS, de Silva R, Rutledge SL, Bohlega S, Ashizawa T, Zoghbi HY (1998) Molecular and clinical studies in SCA-7 define a broad clinical spectrum and the infantile phenotype. Neurology 51:1081–1086
Christensen BC et al (2009) Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet 5:e1000602. doi:10.1371/journal.pgen.1000602
Collins PY et al (2011) Grand challenges in global mental health. Nature 475:27–30. doi:10.1038/475027a
Efstratiadis A (1994) Parental imprinting of autosomal mammalian genes. Curr Opin Genet Dev 4:265–280
Fancy SP, Zhao C, Franklin RJ (2004) Increased expression of Nkx2.2 and Olig2 identifies reactive oligodendrocyte progenitor cells responding to demyelination in the adult CNS. Mol Cell Neurosci 27:247–254 (S1044-7431(04)00159-9 [pii])
Gelfman S, Cohen N, Yearim A, Ast G (2013) DNA-methylation effect on cotranscriptional splicing is dependent on GC architecture of the exon-intron structure. Genome Res 23:789–799. doi:10.1101/gr.143503.112
Genome-wide association study identifies five new schizophrenia loci (2011) Nat Genet 43:969–976. doi:10.1038/ng.940
Greenwood TA et al (2013) Genome-wide linkage analyses of 12 endophenotypes for schizophrenia from the Consortium on the Genetics of Schizophrenia. Am J Psychiatry 170:521–532. doi:10.1176/appi.ajp.2012.12020186
Hamshere ML et al (2013) Genome-wide significant associations in schizophrenia to ITIH3/4, CACNA1C and SDCCAG8, and extensive replication of associations reported by the Schizophrenia PGC. Mol Psychiatry 18:708–712. doi:10.1038/mp.2012.67
Harris EC, Barraclough B (1998) Excess mortality of mental disorder. Br J Psychiatry 173:11–53
Hastie T, Tibshirani R, Friedman J (2001) The elements of statistical learning: data mining, inference, and prediction. Springer, New York
Henckel A, Nakabayashi K, Sanz LA, Feil R, Hata K, Arnaud P (2009) Histone methylation is mechanistically linked to DNA methylation at imprinting control regions in mammals. Hum Mol Genet 18:3375–3383. doi:10.1093/hmg/ddp277
Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis (2013) Lancet 381:1371–1379. doi:10.1016/S0140-6736(12)62129-1
Kandel ER (2012) The molecular biology of memory: cAMP, PKA, CRE, CREB-1, CREB-2, and CPEB Mol. Brain 5:14. doi:10.1186/1756-6606-5-14
Karolchik D et al (2014) The UCSC Genome Browser database: 2014 update. Nucleic Acids Res 42:D764–D770. doi:10.1093/nar/gkt1168
Kenedy AA, Cohen KJ, Loveys DA, Kato GJ, Dang CV (2003) Identification and characterization of the novel centrosome-associated protein CCCAP. Gene 303:35–46 (S0378111902011411 [pii])
Kerkel K et al (2008) Genomic surveys by methylation-sensitive SNP analysis identify sequence-dependent allele-specific DNA methylation. Nat Genet 40:904–908. doi:10.1038/ng.174
Kumar P, Henikoff S, Ng PC (2009) Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 4:1073–1081. doi:10.1038/nprot.2009.86
Liu X, Jian X, Boerwinkle E (2013) dbNSFP v2.0: a database of human non-synonymous SNVs and their functional predictions and annotations. Hum Mutat 34:E2393–E2402. doi:10.1002/humu.22376
Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T (2010) Regulation of alternative splicing by histone modifications. Science 327:996–1000. doi:10.1126/science.1184208
Montminy M (1997) Transcriptional regulation by cyclic AMP. Annu Rev Biochem 66:807–822. doi:10.1146/annurev.biochem.66.1.807
Murray CJ, Lopez AD (1996) Evidence-based health policy—lessons from the Global Burden of Disease Study. Science 274:740–743
Murray RM, Jones PB, Susser E, Van Os J, Cannon M (eds) (2003) The epidemiology of schizophrenia. Cambridge University Press, Cambridge. doi:10.1093/ije/dyh076
Niculescu AB 3rd, Segal DS, Kuczenski R, Barrett T, Hauger RL, Kelsoe JR (2000) Identifying a series of candidate genes for mania and psychosis: a convergent functional genomics approach. Physiol Genomics 4:83–91 (4/1/83 [pii])
Prendergast GC, Ziff EB (1991) Methylation-sensitive sequence-specific DNA binding by the c-Myc basic region. Science 251:186–189
Rakyan VK, Down TA, Balding DJ, Beck S (2011) Epigenome-wide association studies for common human diseases. Nat Rev Genet 12:529–541. doi:10.1038/nrg3000
Ripke S et al (2013) Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet 45:1150–1159. doi:10.1038/ng.2742
Rothbart SB et al (2012) Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation. Nat Struct Mol Biol 19:1155–1160. doi:10.1038/nsmb.2391
Serre D, Lee BH, Ting AH (2010) MBD-isolated genome sequencing provides a high-throughput and comprehensive survey of DNA methylation in the human genome. Nucleic Acids Res 38:391–399. doi:10.1093/nar/gkp992
Shi J et al (2009) Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature 460:753–757. doi:10.1038/nature08192
Shoemaker R, Deng J, Wang W, Zhang K (2010) Allele-specific methylation is prevalent and is contributed by CpG-SNPs in the human genome. Genome Res 20:883–889. doi:10.1101/gr.104695.109
Sullivan PF, Kendler KS, Neale MC (2003) Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch Gen Psychiatry 60:1187–1192. doi:10.1001/archpsyc.60.12.1187
Sullivan PF, Daly MJ, O’Donovan M (2012) Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nat Rev Genet 13:537–551. doi:10.1038/nrg3240
Sutherland JE, Costa M (2003) Epigenetics and the environment. Ann NY Acad Sci 983:151–160
Uhlhaas PJ, Singer W (2010) Abnormal neural oscillations and synchrony in schizophrenia. Nat Rev Neurosci 11:100–113. doi:10.1038/nrn2774
Young AM, Campbell E, Lynch S, Suckling J, Powis SJ (2011) Aberrant NF-kappaB expression in autism spectrum condition: a mechanism for neuroinflammation. Front Psychiatry 2:27. doi:10.3389/fpsyt.2011.00027
Acknowledgments
This study was supported by the National Institute of Mental Health (Grant 1R01MH097283). The present study is part of a larger project entitled ‘A Large-Scale Schizophrenia Association Study in Sweden’ that is supported by Grants from NIMH (MH077139) and the Stanley Medical Research Institute. Institutions involved in this Project are: Karolinska Institute, Icahn School of Medicine at Mount Sinai, University of North Carolina at Chapel Hill, Virginia Commonwealth University, Broad Institute, and the US National Institute of Mental Health. Library construction and next-generation sequencing was performed by EdgeBio Gaithersburg, MD.
Conflict of interest
The authors declare no competing financial interests.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kumar, G., Clark, S.L., McClay, J.L. et al. Refinement of schizophrenia GWAS loci using methylome-wide association data. Hum Genet 134, 77–87 (2015). https://doi.org/10.1007/s00439-014-1494-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-014-1494-5