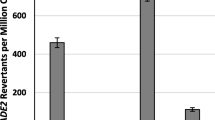
Abstract
The eukaryotic mariner transposons are currently thought to have no sequence specificity for integration other than to insert within a TA contained in a degenerated [TA]1–4 tract, either in vitro or in vivo. We have investigated the properties of a suspected hotspot for the integration of the mariner Mos1 element, namely the Tn9 cat gene that encodes a chloramphenicol acetyl transferase. Using in vitro and bacterial transposition assays, we confirmed that the cat gene is a preferential target for MOS1 integration, whatever its sequence environment, copy number or chromosomal locus. We also observed that its presence increases transposition rates both in vitro and in bacterial assays. The structural and sequence features that constitute the attractiveness of cat were also investigated. We first demonstrated that supercoiling is essential for the cat gene to be a hot spot. In contrast to the situation for Tc1-like elements, DNA curvature and bendability were not found to affect integration target preferences. We found that Mos1 integrations do not occur randomly along the cat gene. All TA dinucleotides that are preferred for integration were found within either TATA or TA×TA motifs. However, these motifs are not sufficient to constitute an attractive dinucleotide, since four TATA and TA×TA sites are cold spots.






Similar content being viewed by others
References
Alton NK, Vapnek D (1979) Nucleotide sequence analysis of the chloramphenicol resistance transposon Tn9. Nature 282:864–869
Atlung T, Christensen BB, Hansen FG (1999) Role of the rom protein in copy number control of plasmid pBR322 at different growth rates in Escherichia coli K-12. Plasmid 41:110–119
Auge-Gouillou C, Hamelin MH, Demattei MV, Periquet M, Bigot Y (2001) The wild-type conformation of the Mos-1 inverted terminal repeats is suboptimal for transposition in bacteria. Mol Genet Genomics 265:51–57
Auge-Gouillou C, Brillet B, Hamelin MH, Bigot Y (2005) Assembly of the mariner Mos1 synaptic complex. Mol Cell Biol 25:2861–2870
Bancroft I, Dean C (1993) Transposition pattern of the maize element Ds in Arabidopsis thaliana. Genetics 134:1221–1229
Benham CJ, Savitt AG, Bauer WR (2002) Extrusion of an imperfect palindrome to a cruciform in superhelical DNA: complete determination of energetics using a statistical mechanical model. J Mol Biol 316:381–563
Consortium TCe (1998) Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282:2012–2018
Demarre G, Guerout AM, Matsumoto-Mashimo C, Rowe-Magnus DA, Marliere P, Mazel D (2005) A new family of mobilizable suicide plasmids based on broad host range R388 plasmid (IncW) and RP4 plasmid (IncPalpha) conjugative machineries and their cognate Escherichia coli host strains. Res Microbiol 156:245–255
Fischer SE, Wienholds E, Plasterk RH (2003) Continuous exchange of sequence information between dispersed Tc1 transposons in the Caenorhabditis elegans genome. Genetics 164:127–134
Granger L, Martin E, Segalat L (2004) Mos as a tool for genome-wide insertional mutagenesis in Caenorhabditis elegans: results of a pilot study. Nucl Acids Res 32:e117
Groenen MA, Kokke M, van de Putte P (1986) Transposition of mini-Mu containing only one of the ends of bacteriophage Mu. EMBO J 5:3687–3690
Hackett CS, Geurts AM, Hackett PB (2007) Predicting preferential DNA vector insertion sites: implications for functional genomics and gene therapy. Genome Biol 8(Suppl 1):S12
Halaimia-Toumi N, Casse N, Demattei MV, Renault S, Pradier E, Bigot Y, Laulier M (2004) The GC-rich transposon Bytmar1 from the deep-sea hydrothermal crab, Bythograea thermydron, may encode three transposase isoforms from a single ORF. J Mol Evol 59:747–760
Horie K, Yusa K, Yae K, Odajima J, Fischer SE, Keng VW, Hayakawa T, Mizuno S, Kondoh G, Ijiri T, Matsuda Y, Plasterk RH, Takeda J (2003) Characterization of Slee** Beauty transposition and its application to genetic screening in mice. Mol Cell Biol 23:9189–9207
Huang J, Zhang Q, Schlick T (2003) Effect of DNA superhelicity and bound proteins on mechanistic aspects of the Hin-mediated and Fis-enhanced inversion. Biophys J 85:804–817
Kado CI, Liu ST (1981) Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol 145:1365–1373
Ketting RF, Fischer SE, Plasterk RH (1997) Target choice determinants of the Tc1 transposon of Caenorhabditis elegans. Nucl Acids Res 25:4041–4047
Lipkow K, Buisine N, Chalmers R (2004) Promiscuous target interactions in the mariner transposon Himar1. J Biol Chem 279:48569–48575
Liu D, Bischerour J, Siddique A, Buisine N, Bigot Y, Chalmers R (2007) The human SETMAR protein preserves most of the activities of the ancestral Hsmar1 transposase. Mol Cell Biol 27:1125–1132
Lohe AR, Hartl DL (2002) Efficient mobilization of mariner in vivo requires multiple internal sequences. Genetics 160:519–526
Neiditch MB, Lee GS, Landree MA, Roth DB (2001) RAG transposase can capture and commit to target DNA before or after donor cleavage. Mol Cell Biol 21:4302–4310
Niehus E, Cheng E, Tan M (2008) DNA supercoiling-dependent gene regulation in Chlamydia. J Bacteriol 190:6419–6427
Petit A, Sirot DL, Chanal CM, Sirot JL, Labia R, Gerbaud G, Cluzel RA (1988) Novel plasmid-mediated beta-lactamase in clinical isolates of Klebsiella pneumoniae more resistant to ceftazidime than to other broad-spectrum cephalosporin. Antimicrob Agents Chemother 32:626–630
Pogliano J, Ho TQ, Zhong Z, Helinski DR (2001) Multicopy plasmids are clustered and localized in Escherichia coli. Proc Natl Acad Sci USA 98:4486–4491
Riley M, Abe T, Arnaud MB, Berlyn MK, Blattner FR, Chaudhuri RR, Glasner JD, Horiuchi T, Keseler IM, Kosuge T, Mori H, Perna NT, Plunkett G 3rd, Rudd KE, Serres MH, Thomas GH, Thomson NR, Wishart D, Wanner BL (2006) Escherichia coli K-12: a cooperatively developed annotation snapshot-2005. Nucl Acids Res 34:1–9
Rouleux-Bonnin F, Bigot S, Bigot Y (2004) Structural and transcriptional features of Bombus terrestris satellite DNA and their potential involvement in the differentiation process. Genome 47:877–888
Sakai J, Kleckner N (1997) The Tn10 synaptic complex can capture a target DNA only after transposon excision. Cell 89:205–214
Sambrook J, Russel D (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, New York
Tower J, Karpen GH, Craig N, Spradling AC (1993) Preferential transposition of Drosophila P elements to nearby chromosomal sites. Genetics 133:347–359
van Luenen HG, Plasterk RH (1994) Target site choice of the related transposable elements Tc1 and Tc3 of Caenorhabditis elegans. Nucl Acids Res 22:262–269
Viglasky V, Danko P (2007) Intercalators: contra cruciform extrusion in DNA. Anal Biochem 360:7–13
Vos JC, De Baere I, Plasterk RH (1996) Transposase is the only nematode protein required for in vitro transposition of Tc1. Genes Dev 10:755–761
Yao S, Helinski DR, Toukdarian A (2007) Localization of the naturally occurring plasmid ColE1 at the cell pole. J Bacteriol 189:1946–1953
Weitao T, Nordström K, Dasgupta S (2000) Escherichia coli cell cycle control genes affect chromosome superhelicity. EMBO Rep 1:494–499
Acknowledgments
This work was supported by the University of François Rabelais of Tours, and funded by grants from the European Commission (Project SyntheGeneDelivery, No 018716), the CNRS, the French Ministère de l’Education Nationale, de la Recherche et de la Technologie (MENRT), the Association Française contre la Myopathie, the Groupement de Recherche CNRS 2157. G. Crénès holds a doctoral fellowship from the European Commission, and S. Dion from the French MENRT. We would like to thank Dr. M. Chandler for providing chloramphenicol-mediated resistance strains, Dr. G. Demarre for providing the pSW29 plasmid, and Dr. L. Segalat providing the sequences of insertion in C. elegans rDNA. The English text has been revised by Dr. M. Ghosh.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by D. Andersson.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Crénès, G., Ivo, D., Hérisson, J. et al. The bacterial Tn9 chloramphenicol resistance gene: an attractive DNA segment for Mos1 mariner insertions. Mol Genet Genomics 281, 315–328 (2009). https://doi.org/10.1007/s00438-008-0414-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-008-0414-6