Abstract
Main conclusion
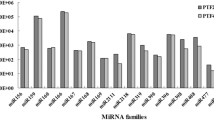
Using a genome-wide analysis of miRNAs in ‘Yali’ pear (Pyrus bretschneideri) via the next-generation high-throughput sequencing of small RNAs with a bioinformatics analysis, we found that pbr-miR156, pbr-miR164, pbr-miR399, and pbr-miR482 and their target genes function in viral defense in ‘Duli’ and ‘Hongbaoshi’. pbr-miR160, pbr-miR168, pbr-miR171, and pbr-miR319 and their targets function in auxin signaling pathways in ‘Zhongai 4’ and ‘Zhongai 5’.
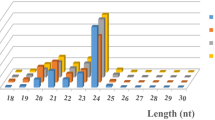
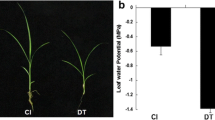
Successful fruit production in pear (Pyrus spp.) depends on the use of optimal combinations of rootstocks and scions. Deciphering plant–pathogen defense mechanisms and hormone signaling pathways is an important step towards develo** pear rootstocks and varieties with improved qualities. In the current study, we combined next-generation sequencing of small RNAs with a bioinformatics analysis to systematically identify and characterize 298 miRNAs in the pear scion cultivar ‘Yali’ (Pyrus bretschneideri). We also analyzed miRNAs in three rootstock varieties (‘Duli’, ‘Zhongai 4’, and ‘Zhongai 5’) and one scion cultivar (‘Hongbaoshi’). We found that pbr-miR156, pbr-miR164, pbr-miR399, and pbr-miR482 are induced following infection with the pear virus Apple stem pitting virus (ASPV), and identified their target genes (pbRPS6, pbNAC, pbTLR, and pbRX-CC, respectively), which participate in viral defense pathways in ‘Duli’ and ‘Hongbaoshi’. Furthermore, we identified pbr-miR160, pbr-miR168, pbr-miR171, and pbr-miR319, and found that the production of these miRNAs was suppressed under low levels of synthetic auxin. The targets of these miRNAs (pbARF, pbAEC, pbSCL, and pbTCP4) respond to auxin signaling pathways in ‘Zhongai 4’ and ‘Zhongai 5’. Our results lay the foundation for breeding improved pear cultivars.







Similar content being viewed by others
Abbreviations
- R gene:
-
Resistance gene
- TLRs:
-
Toll-like receptors
- ARF:
-
Auxin response factor
- AEC:
-
Auxin efflux carrier
- ASGV:
-
Apple stem grooving virus
- ASPV:
-
Apple stem pitting virus
- ACLSV:
-
Apple chlorotic leaf spot virus
- NAA:
-
1-naphthaleneacetic acid; synthetic auxin
- SCL:
-
Scarecrow-like
References
Allen E, **e Z, Gustafson AM, Carrington JC (2005) microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121(2):207–221. https://doi.org/10.1016/j.cell.2005.04.004
Axtell MJ (2013) Classification and comparison of small RNAs from plants. Annu Rev Plant Biol 64:137–159. https://doi.org/10.1146/annurev-arplant-050312-120043
Axtell MJ, Meyers BC (2018) Revisiting criteria for plant microRNA annotation in the era of big data. Plant Cell 30(2):272–284. https://doi.org/10.1105/tpc.17.00851
Baker CC, Sieber P, Wellmer F, Meyerowitz EM (2005) The early extra petals1 mutant uncovers a role for microRNA miR164c in regulating petal number in Arabidopsis. Curr Biol 15(4):303–315. https://doi.org/10.1016/j.cub.2005.02.017
Barakat A, Sriram A, Park J, Zhebentyayeva T, Main D, Abbott A (2012) Genome wide identification of chilling responsive microRNAs in Prunus persica. BMC Genomics 13:481. https://doi.org/10.1186/1471-2164-13-481
Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136(2):215–233. https://doi.org/10.1016/j.cell.2009.01.002
Baulcombe D (2004) RNA silencing in plants. Nature 431(7006):356–363. https://doi.org/10.1038/nature02874
Belli Kullan J, Lopes Paim Pinto D, Bertolini E, Fasoli M, Zenoni S, Tornielli GB, Pezzotti M, Meyers BC, Farina L, Pe ME, Mica E (2015) miRVine: a microRNA expression atlas of grapevine based on small RNA sequencing. BMC Genomics 16:393. https://doi.org/10.1186/s12864-015-1610-5
Bent AF, Mackey D (2007) Elicitors, effectors, and R genes: the new paradigm and a lifetime supply of questions. Annu Rev Phytopathol 45:399–436. https://doi.org/10.1146/annurev.phyto.45.062806.094427
Carrera M, Gomez-Aparasi J (1998) Rootstock influence on the performance of the peach variety ‘Catherine’. Fourth Int Peach Symp 1–2(465):573–577. https://doi.org/10.17660/ActaHortic.1998.465.71
Casey LW, Lavrencic P, Bentham AR, Cesari S, Ericsson DJ, Croll T, Turk D, Anderson PA, Mark AE, Dodds PN, Mobli M, Kobe B, Williams SJ (2016) The CC domain structure from the wheat stem rust resistance protein Sr33 challenges paradigms for dimerization in plant NLR proteins. Proc Natl Acad Sci USA 113(45):12856–12861. https://doi.org/10.1073/pnas.1609922113
Chellappan P, ** H (2009) Discovery of plant microRNAs and short-interfering RNAs by deep parallel sequencing. Methods Mol Biol 495:121–132. https://doi.org/10.1007/978-1-59745-477-3_11
Chen XM (2004) A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303(5666):2022–2025. https://doi.org/10.1126/science.1088060
Chisholm ST, Coaker G, Day B, Staskawicz BJ (2006) Host-microbe interactions: sha** the evolution of the plant immune response. Cell 124(4):803–814. https://doi.org/10.1016/j.cell.2006.02.008
Ditengou FA, Teale WD, Kochersperger P, Flittner KA, Kneuper I, van der Graaff E, Nziengui H, Pinosa F, Li X, Nitschke R, Laux T, Palme K (2008) Mechanical induction of lateral root initiation in Arabidopsis thaliana. Proc Natl Acad Sci USA 105(48):18818–18823. https://doi.org/10.1073/pnas.0807814105
Ditengou FA, Gomes D, Nziengui H, Kochersperger P, Lasok H, Medeiros V, Paponov IA, Nagy SK, Nadai TV, Meszaros T, Barnabas B, Ditengou BI, Rapp K, Qi L, Li X, Becker C, Li C, Doczi R, Palme K (2018) Characterization of auxin transporter PIN6 plasma membrane targeting reveals a function for PIN6 in plant bolting. New Phytol 217(4):1610–1624. https://doi.org/10.1111/nph.14923
Dodds PN, Rathjen JP (2010) Plant immunity: towards an integrated view of plant-pathogen interactions. Nature Rev Genetics 11(8):539–548. https://doi.org/10.1038/nrg2812
Fang Y, Liao K, Du H, Xu Y, Song H, Li X, **ong L (2015) A stress-responsive NAC transcription factor SNAC3 confers heat and drought tolerance through modulation of reactive oxygen species in rice. J Exp Bot 66(21):6803–6817. https://doi.org/10.1093/jxb/erv386
Feng H, Duan X, Zhang Q, Li X, Wang B, Huang L, Wang X, Kang Z (2014) The target gene of tae-miR164, a novel NAC transcription factor from the NAM subfamily, negatively regulates resistance of wheat to stripe rust. Mol Plant Pathol 15(3):284–296. https://doi.org/10.1111/mpp.12089
Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jurgens G (2003) Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426(6963):147–153. https://doi.org/10.1038/nature02085
Gao Z, Shi T, Luo X, Zhang Z, Zhuang W, Wang L (2012) High-throughput sequencing of small RNAs and analysis of differentially expressed microRNAs associated with pistil development in Japanese apricot. BMC Genomics 13:371. https://doi.org/10.1186/1471-2164-13-371
Giorgi M, Capocasa F, Scalzo J, Murri G, Battino M, Mezzetti B (2005) The rootstock effects on plant adaptability, production, fruit quality, and nutrition in the peach (cv. ‘Suncrest’). Sci Hortic-Amsterdam 107(1):36–42. https://doi.org/10.1016/j.scienta.2005.06.003
Gray WM (2004) Hormonal regulation of plant growth and development. PLoS Biol 2(9):E311. https://doi.org/10.1371/journal.pbio.0020311
Guan QM, Lu XY, Zeng HT, Zhang YY, Zhu JH (2013) Heat stress induction of miR398 triggers a regulatory loop that is critical for thermotolerance in Arabidopsis. Plant J 74(5):840–851. https://doi.org/10.1111/tpj.12169
Guilfoyle TJ, Hagen G (2007) Auxin response factors. Curr Opin Plant Biol 10(5):453–460. https://doi.org/10.1016/j.pbi.2007.08.014
Guo R, Chen X, Lin Y, Xu X, Thu MK, Lai Z (2015) Identification of novel and conserved miRNAs in leaves of in vitro grown Citrus reticulata “Lugan” plantlets by Solexa Sequencing. Front Plant Sci 6:1212. https://doi.org/10.3389/fpls.2015.01212
Gutierrez L, Bussell JD, Pacurar DI, Schwambach J, Pacurar M, Bellini C (2009) Phenotypic plasticity of adventitious rooting in Arabidopsis is controlled by complex regulation of AUXIN RESPONSE FACTOR transcripts and microRNA abundance. Plant Cell 21(10):3119–3132. https://doi.org/10.1105/tpc.108.064758
Huang W, Peng S, **an Z, Lin D, Hu G, Yang L, Ren M, Li Z (2017) Overexpression of a tomato miR171 target gene SlGRAS24 impacts multiple agronomical traits via regulating gibberellin and auxin homeostasis. Plant Biotechnol J 15(4):472–488. https://doi.org/10.1111/pbi.12646
Jones JD, Dangl JL (2006) The plant immune system. Nature 444(7117):323–329. https://doi.org/10.1038/nature05286
Karimi M, Ghazanfari F, Fadaei A, Ahmadi L, Shiran B, Rabei M, Fallahi H (2016) The small-RNA profiles of almond (Prunus dulcis Mill.) reproductive tissues in response to cold stress. PLoS One 11(6):e0156519. https://doi.org/10.1371/journal.pone.0156519
Kidner CA, Martienssen RA (2005) The developmental role of microRNA in plants. Curr Opin Plant Biol 8(1):38–44. https://doi.org/10.1016/j.pbi.2004.11.008
Kim SH, Il Kwon S, Saha D, Anyanwu NC, Gassmann W (2009) Resistance to the Pseudomonas syringae effector HopA1 is governed by the TIR-NBS-LRR protein RPS6 and is enhanced by mutations in SRFR1. Plant Physiol 150(4):1723–1732. https://doi.org/10.1104/pp.109.139238
Koganezawa H, Yanase H (1990) A new type of elongated virus isolated from apple-trees containing the stem pitting agent. Plant Dis 74(8):610–614. https://doi.org/10.1094/Pd-74-0610
Koyama T, Sato F, Ohme-Takagi M (2017) Roles of miR319 and TCP transcription factors in leaf development. Plant Physiol 175(2):874–885. https://doi.org/10.1104/pp.17.00732
Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T (2001) Identification of novel genes coding for small expressed RNAs. Science 294(5543):853–858. https://doi.org/10.1126/science.1064921
Lau NC, Lim LP, Weinstein EG, Bartel DP (2001) An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294(5543):858–862. https://doi.org/10.1126/science.1065062
Lee RC, Ambros V (2001) An extensive class of small RNAs in Caenorhabditis elegans. Science 294(5543):862–864. https://doi.org/10.1126/science.1065329
Lee RC, Feinbaum RL, Ambros V (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75(5):843–854
Li MF, Li XF, Han ZH, Shu HR, Li T (2009) Molecular analysis of two Chinese pear (Pyrus bretschneideri Rehd.) spontaneous self-compatible mutants, Yan Zhuang and ** Zhui. Plant Biol (Stuttg) 11(5):774–783. https://doi.org/10.1111/j.1438-8677.2008.00180.x
Liu Y, Ke L, Wu G, Xu Y, Wu X, **a R, Deng X, Xu Q (2017) miR3954 is a trigger of phasiRNAs that affects flowering time in citrus. Plant J 92(2):263–275. https://doi.org/10.1111/tpj.13650
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25(4):402–408. https://doi.org/10.1006/meth.2001.1262
Llave C, **e ZX, Kasschau KD, Carrington JC (2002) Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297(5589):2053–2056. https://doi.org/10.1126/science.1076311
Ma C, Lu Y, Bai S, Zhang W, Duan X, Meng D, Wang Z, Wang A, Zhou Z, Li T (2014) Cloning and characterization of miRNAs and their targets, including a novel miRNA-targeted NBS-LRR protein class gene in apple (Golden Delicious). Mol Plant 7(1):218–230. https://doi.org/10.1093/mp/sst101
Mao Y, Wu F, Yu X, Bai J, Zhong W, He Y (2014) microRNA319a-targeted Brassica rapa ssp. pekinensis TCP genes modulate head shape in chinese cabbage by differential cell division arrest in leaf regions. Plant Physiol 164(2):710–720. https://doi.org/10.1104/pp.113.228007
Martelli GP, Adams MJ, Kreuze JF, Dolja VV (2007) Family Flexiviridae: a case study in virion and genome plasticity. Annu Rev Phytopathol 45:73–100. https://doi.org/10.1146/annurev.phyto.45.062806.094401
Millar AA, Gubler F (2005) The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell 17(3):705–721. https://doi.org/10.1105/tpc.104.027920
Nandety RS, Caplan JL, Cavanaugh K, Perroud B, Wroblewski T, Michelmore RW, Meyers BC (2013) The role of TIR-NBS and TIR-X proteins in plant basal defense responses. Plant Physiol 162(3):1459–1472. https://doi.org/10.1104/pp.113.219162
Pagliarani C, Vitali M, Ferrero M, Vitulo N, Incarbone M, Lovisolo C, Valle G, Schubert A (2017) The accumulation of miRNAs differentially modulated by drought stress is affected by grafting in grapevine. Plant Physiol 173(4):2180–2195. https://doi.org/10.1104/pp.16.01119
Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D (2003) Control of leaf morphogenesis by microRNAs. Nature 425(6955):257–263. https://doi.org/10.1038/nature01958
Pantaleo V, Szittya G, Moxon S, Miozzi L, Moulton V, Dalmay T, Burgyan J (2010) Identification of grapevine microRNAs and their targets using high-throughput sequencing and degradome analysis. Plant Journal 62(6):960–976. https://doi.org/10.1111/j.1365-313X.2010.04208.x
Radhakrishnan GK, Splitter GA (2010) Biochemical and functional analysis of TIR domain containing protein from Brucella melitensis. Biochem Biophys Res Commun 397(1):59–63. https://doi.org/10.1016/j.bbrc.2010.05.056
Rairdan GJ, Collier SM, Sacco MA, Baldwin TT, Boettrich T, Moffett P (2008) The coiled-coil and nucleotide binding domains of the potato Rx disease resistance protein function in pathogen recognition and signaling. Plant Cell 20(3):739–751. https://doi.org/10.1105/tpc.107.056036
Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP (2002) Prediction of plant microRNA targets. Cell 110(4):513–520. https://doi.org/10.1016/S0092-8674(02)00863-2
Sabatini S, Beis D, Wolkenfelt H, Murfett J, Guilfoyle T, Malamy J, Benfey P, Leyser O, Bechtold N, Weisbeek P, Scheres B (1999) An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99(5):463–472
Santner A, Calderon-Villalobos LIA, Estelle M (2009) Plant hormones are versatile chemical regulators of plant growth. Nat Chem Biol 5(5):301–307. https://doi.org/10.1038/nchembio.165
Schommer C, Debernardi JM, Bresso EG, Rodriguez RE, Palatnik JF (2014) Repression of cell proliferation by miR319-regulated TCP4. Mol Plant 7(10):1533–1544. https://doi.org/10.1093/mp/ssu084
Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D (2005) Specific effects of microRNAs on the plant transcriptome. Dev Cell 8(4):517–527. https://doi.org/10.1016/j.devcel.2005.01.018
Schwarz K, Jelkmann W (1998) Detection and characterization of European apple stem pitting virus sources from apple and pear by PCR and partial sequence analysis. Acta Hortic 472:75–85. https://doi.org/10.17660/ActaHortic.1998.472.6
Su H, Zhang S, Yuan X, Chen C, Wang XF, Hao YJ (2013) Genome-wide analysis and identification of stress-responsive genes of the NAM-ATAF1, 2-CUC2 transcription factor family in apple. Plant Physiol Biochem 71:11–21. https://doi.org/10.1016/j.plaphy.2013.06.022
Suer S, Agusti J, Sanchez P, Schwarz M, Greb T (2011) WOX4 imparts auxin responsiveness to cambium cells in Arabidopsis. Plant Cell 23(9):3247–3259. https://doi.org/10.1105/tpc.111.087874
Sunkar R, Zhu JK (2004) Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16(8):2001–2019. https://doi.org/10.1105/tpc.104.022830
Torto TA, Rauser L, Kamoun S (2002) The pipg1 gene of the oomycete Phytophthora infestans encodes a fungal-like endopolygalacturonase. Curr Genet 40(6):385–390. https://doi.org/10.1007/s00294-002-0272-4
Varkonyi-Gasic E, Gould N, Sandanayaka M, Sutherland P, MacDiarmid RM (2010) Characterisation of microRNAs from apple (Malus domestica ‘Royal Gala’) vascular tissue and phloem sap. BMC Plant Biol 10:159. https://doi.org/10.1186/1471-2229-10-159
Vaucheret H (2006) Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev 20(7):759–771. https://doi.org/10.1101/gad.1410506
Voinnet O (2009) Origin, biogenesis, and activity of plant microRNAs. Cell 136(4):669–687. https://doi.org/10.1016/j.cell.2009.01.046
Wang Q, Cuellar WJ, Rajamaki ML, Hirata Y, Valkonen JP (2008) Combined thermotherapy and cryotherapy for efficient virus eradication: relation of virus distribution, subcellular changes, cell survival and viral RNA degradation in shoot tips. Mol Plant Pathol 9(2):237–250. https://doi.org/10.1111/j.1364-3703.2007.00456.x
Wang B, Wang J, Wang C, Shen W, Jia H, Zhu X, Li X (2016) Study on expression modes and cleavage role of miR156b/c/d and its target gene Vv-SPL9 during the whole growth stage of grapevine. J Hered 107(7):626–634. https://doi.org/10.1093/jhered/esw030
Wang M, Sun X, Wang C, Cui L, Chen L, Zhang C, Shangguan L, Fang J (2017) Characterization of miR061 and its target genes in grapevine responding to exogenous gibberellic acid. Funct Integr Genomics 17(5):537–549. https://doi.org/10.1007/s10142-017-0554-z
Yang L, Mu X, Liu C, Cai J, Shi K, Zhu W, Yang Q (2015) Overexpression of potato miR482e enhanced plant sensitivity to Verticillium dahliae infection. J Integr Plant Biol 57(12):1078–1088. https://doi.org/10.1111/jipb.12348
Yao JL, Xu J, Cornille A, Tomes S, Karunairetnam S, Luo Z, Bassett H, Whitworth C, Rees-George J, Ranatunga C, Snirc A, Crowhurst R, de Silva N, Warren B, Deng C, Kumar S, Chagne D, Bus VG, Volz RK, Rikkerink EH, Gardiner SE, Giraud T, MacDiarmid R, Gleave AP (2015) A microRNA allele that emerged prior to apple domestication may underlie fruit size evolution. Plant J 84(2):417–427. https://doi.org/10.1111/tpj.13021
Yu X, Hou Y, Chen W, Wang S, Wang P, Qu S (2017) Malus hupehensis miR168 targets to ARGONAUTE1 and contributes to the resistance against Botryosphaeria dothidea infection by altering defense responses. Plant Cell Physiol 58(9):1541–1557. https://doi.org/10.1093/pcp/pcx080
Zhang CH, Zhang BB, Ma RJ, Yu ML, Guo SL, Guo L (2015) Isolation and expression analysis of four HD-ZIP III family genes targeted by microRNA166 in peach. Genet Mol Res 14(4):14151–14161. https://doi.org/10.4238/2015.October.29.37
Zhang C, Zhang B, Ma R, Yu M, Guo S, Guo L, Korir NK (2016a) Identification of known and novel microRNAs and their targets in peach (Prunus persica) fruit by high-throughput sequencing. PLoS One 11(7):e0159253. https://doi.org/10.1371/journal.pone.0159253
Zhang X, Wang W, Wang M, Zhang HY, Liu JH (2016b) The miR396b of Poncirus trifoliata functions in cold tolerance by regulating ACC oxidase gene expression and modulating ethylene-polyamine homeostasis. Plant Cell Physiol 57(9):1865–1878. https://doi.org/10.1093/pcp/pcw108
Zhang QL, Li Y, Zhang Y, Wu C, Wang S, Hao L, Wang S, Li T (2017) Md-miR156ab and Md-miR395 target WRKY transcription factors to influence apple resistance to leaf spot disease. Front Plant Sci 8:526. https://doi.org/10.3389/fpls.2017.00526
Zhu QH, Fan L, Liu Y, Xu H, Llewellyn D, Wilson I (2013) miR482 regulation of NBS-LRR defense genes during fungal pathogen infection in cotton. PLoS One 8(12):e84390. https://doi.org/10.1371/journal.pone.0084390
Acknowledgements
This study was financially supported by the earmarked fund for China Pear Modern Agro-industry Technology Research System CARS 28-08.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. S1
Characteristic fold-back RNA secondary structure of 21 novel miRNAs. The hairpin structure precursors of 21 potential novel miRNAs identified in a small RNA library constructed from ‘Yali’ leaves. Highlighted in red are the novel miRNA sequences and in blue are the miRNA* sequences (PDF 897 kb)
Supplementary Fig. S2
Conserved domains of pbr-miR156 (a), pbr-miR164 (b), pbr-miR399 (c), and pbr-miR482 (d) target genes pbRPS6, pbNAC, pbTLR, and pbRX-CC, respectively (JPEG 1581 kb)
Supplementary Fig. S3
Conserved domains of pbr-miR160 (a), pbr-miR168 (b), pbr-miR171 (c), and pbr-miR319 (d) target genes pbARF, pbAEC, pbSCL, and pbTCP4, respectively (JPEG 1617 kb)
Supplementary Fig. S4
Diagram of multiple PCR analysis of three viruses (ASGV, ASPV, and ACLSV) (PNG 191 kb)
Rights and permissions
About this article
Cite this article
Zhang, Q., Zhang, Y., Wang, S. et al. Characterization of genome-wide microRNAs and their roles in development and biotic stress in pear. Planta 249, 693–707 (2019). https://doi.org/10.1007/s00425-018-3027-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-018-3027-2