Abstract
Main conclusion
UV-B-caused DNA hypomethylation and UV-B-mediated epigenetic activation of additional WRKY-binding site(s) in the DBR2 promoter may contribute to the overexpression of the DBR2 gene in Artemisia annua.
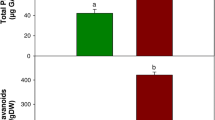
DNA methylation is one of the key mechanisms behind stress-induced transcriptional switch off/on. Here, we evaluate the DNA methylation level in response to UV-B radiation in Artemisia annua which produces artemisinin, a sesquiterpene that has been recommended by WHO for the frontline treatment of malaria. However, the drug is facing serious shortage due to its low concentration in plants. UV-B treatment (3 h) enhanced artemisinin concentration up to 1.91-fold as compared to control. A key regulatory gene of artemisinin biosynthesis, DBR2 was upregulated under UV-B. This study presents observations regarding contributions of DNA methylation to the gene regulation using DBR2 as an example. Restriction digestion of genomic DNA by isoschizomers (MspI and HpaII) suggested UV-B involvement in DNA hypomethylation in A. annua. The global level of DNA methylation (R) was 3.4 and 5.9 % for UV-B treated and control plants, respectively, attesting hypomethylation of DNA in response to UV-B. Further bisulfite sequencing PCR showed demethylation at two CHG sites in 18S rRNA gene. Similarly, bisulfite sequencing of promoter region of DBR2 has demonstrated demethylation at 4 CG-, 4 CHH- and 2 CHG-sites. In silico analysis revealed UV-B-mediated demethylation at seven putative transcription factor binding sites including WRKY, which are positive regulators of artemisinin biosynthesis. UV-B treatment has resulted in activation of additional WRKY-binding site in UV-B-treated plants compared with single active WRKY-binding site in control and this could be the probable reason for overexpression of DBR2. It is suggested that DNA demethylation is an important epigenetic response to UV-B radiation in A. annua that surely will provide new horizons to further elucidate the mechanistic evidence of plant’s responses to UV-B radiation.








Similar content being viewed by others
References
Ahlawat S, Saxena P, Alam P, Wajid S, Abdin MZ (2014) Modulation of artemisinin biosynthesis by elicitors, inhibitor, and precursor in hairy root cultures of Artemisia annua L. J Plant Interact 9(1):811–824. doi:10.1080/17429145.2014.949885
Aina R, Sgorbati S, Santagostino A, Labra M, Ghiani A, Citterio S (2004) Specific hypomethylation of DNA is induced by heavy metals in white clover and industrial hemp. Physiol Plant 121(3):472–480
Akimoto K, Katakami H, Kim HJ, Ogawa E, Sano CM, Wada Y, Sano H (2007) Epigenetic inheritance in rice plants. Ann Bot 100:205–217. doi:10.1093/aob/mcm110
Baker SS, Wilhelm KS, Thomashow MF (1994) The 5′-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol Biol 24(5):701–713
Barciszewska MZ, Barciszewska AM, Rattan SI (2007) TLC-based detection of methylated cytosine: application to aging epigenetics. Biogerontology 8:673–678
Chen H, Lai Z, Shi J, **ao Y, Chen Z, Xu X (2010) Roles of Arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress. BMC Plant Biol 10:281. doi:10.1186/1471-2229-10-281
Deaton AM, Bird A (2011) CpG islands and the regulation of transcription. Genes Dev 25(10):1010–1022
Dhar MK, Vishal P, Sharma R, Kaul S (2014) Epigenetic dynamics: role of epimarks and underlying machinery in plants exposed to abiotic stress. Journal of Genomics, vol 2014, Hindawi Publishing Corporation International, Article ID 187146, p 10. doi:10.1155/2014/187146
Dyachenko O, Zakharchenko N, Shevchuk T, Bohnert HJ, Cushman JC, Buryanov YI (2006) Effect of hypermethylation of CCWGG sequences in DNA of Mesembryanthemum crystallinum plants on their adaptation to salt stress. Biochemistry (Moscow) 71(4):461–465
Fang H, Liu X, Thorn G, Duan J, Tian L (2014) Expression analysis of histone acetyltransferases in rice under drought stress. Biochem Biophys Res Commun 443:400–405. doi:10.1016/j.bbrc.2013.11.102
Galaud JP, Gaspar T, Boyer N (1993) Inhibition of internode growth due to mechanical stress in Bryonia dioica: relationship between changes in DNA methylation and ethylene metabolism. Physiol Plant 87:25–30
Hasbun R, Valledor L, Rodrıguez JL, Santamaria E, Rıos D, Sanchez M, Cañal MJ, Rodríguez R (2008) HPCE quantification of 5-methyl-20-deoxycytidine in genomic DNA: methodological optimization for chestnut and other woody species. Plant Physiol Biochem 46:815–822
Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27:297–300
** H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C (2000) Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J 19:6150–6161
Khanuja SPS, Shasany AK, Darokar MP, Kumar S (1999) Rapid isolation of DNA from dry and fresh samples of plants producing large amounts of secondary metabolites and essential oils. Plant Mol Bio Rep 17:1–7
Kumari R, Yadav G, Sharma V, Sharma V, Kumar S (2013) Cytosine hypomethylation at CHG and CHH sites in the pleiotropic mutants of Mendelian inheritance in Catharanthus roseus. J Genet 92(3):499–511
Labra M, Grassi F, Imazio S, Di Fabio T, Citterio S, Sgorbati S, Agradi E (2004) Genetic and DNA-methylation changes induced by potassium dichromate in Brassica napus L. Chemosphere 54(8):1049–1058
Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30:325–327
Li LC, Dahiya R (2002) MethPrimer: designing primers for methylation PCRs. Bioinformatics 18:1427e1431
Ma DD, Pu G, Lei C, Ma L, Wang H, Guo Y, Chen J, Du Z, Wang H, Li G, Ye H, Liu B (2009) Isolation and characterization of AaWRKY1, an Artemisia annua transcription factor that regulates the amorpha-4, 11-diene synthase gene, a key gene of artemisinin biosynthesis. Plant Cell Physiol 50:2146–2161
Medvedeva YA, Khamis AM, Kulakovskiy IV, Ba-Alawi W, Bhuyan, Kawaji H, Lassmann T, Harbers M, Forrest AR, Bajic VB, FANTOM consortium (2013) Effects of cytosine methylation on transcription factor binding sites. BMC Genomics 15:119. doi:10.1186/1471-2164-15-119
Michalak M, Barciszewska MZ, Barciszewski J, Plitta BP, Chmielarz P (2013) Global changes in DNA methylation in seeds and seedlings of Pyrus communis after seed desiccation and storage. PLoS One 8(8):e70693. doi:10.1371/journal.pone.0070693
Mirouze M, Paszkowski J (2011) Epigenetic contribution to stress adaptation in plants. Curr Opin Plant Biol 14:267–274
Ohlsson AB, Segerfeldt P, Lindstrom A, Borg-Karlson AK, Berglund T (2013) UV-B exposure of indoor-grown Picea abies seedlings causes an epigenetic effect and selective emission of terpenes. Z Naturforsch C 68:139–147. doi:10.5560/ZNC.2013.68c0139
Palacios D, Summerbell D, Rigby PWJ, Boyes J (2010) Interplay between DNA methylation and transcription factor availability: implications for developmental activation of the mouse Myogenin gene. Mol Cell Biol 30:3805–3815. doi:10.1128/MCB.00050-10
Pandey N, Pandey-Rai S (2014) Short-term UV-B radiation–mediated transcriptional responses and altered secondary metabolism of in vitro propagated plantlets of Artemisia annua L. Plant Cell Tiss Organ Cult 116:371–385. doi:10.1007/s11240-013-0413-0
Pandey SP, Somssich IE (2009) The role of WRKY transcription factors in plant immunity. Plant Physiol 150:1648–1655. doi:10.1104/pp.109.138990
Peng H, Zhang J (2009) Plant genomic DNA methylation in response to stresses: potential applications and challenges in plant breeding. Prog Nat Sci 19:1037–1045
Sarvestani R, Peyghambary SA, Abbasi A (2014) Isolation and characterization of DBR2 gene promoter from Iranian Artemisia annua. J Agr Sci Tech 16:191–202
Steward N, Kusano T, Sano H (2000) Expression of ZmMET1, a gene encoding a DNA methyltransferase from maize, is associated not only with DNA replication in actively proliferating cells, but also with altered DNA methylation status in cold-stressed quiescent cells. Nucleic Acids Res 28(17):3250–3259
Wada Y, Miyamoto K, Kusano T, Sano H (2004) Association between up-regulation of stress-responsive genes and hypomethylation of genomic DNA in tobacco plants. Mol Genet Genomics 271:658–666
Wang YY, Shu Y, **ao YF, Wang Q, Kanekura T, Li YP, Wang JC, Zhao M, Qian** L, **ao R (2014) Hypomethylation and overexpression of ITGAL (CD11a) in CD4+ T cells in systemic sclerosis. Clin Epigenetics 6(1):25. doi:10.1186/1868-7083-6-25
Yang JL, Liu LW, Gong YQ, Huang DQ, Wang F, He LL (2007) Analysis of genomic DNA methylation level in radish under cadmium stress by methylation sensitive amplified polymorphism technique. J Plant Physiol Mol Biol 33(3):219–226
Yang K, Monafared RS, Wang H, Lundgren A, Brodelius PE (2015) The activity of the artemisinic aldehyde Δ11(13) reductase promoter is important for artemisinin yield in different chemotypes of Artemisia annua L. Plant Mol Biol. doi:10.1007/s11103-015-0284-3
Yuan Y, Liu W, Zhang Q, **ang L, Liu X, Chen M, Lin Z, Wang Q, Liao Z (2015) Overexpression of artemisinic aldehyde Δ11 (13) reductase gene-enhanced artemisinin and its relative metabolite biosynthesis in transgenic Artemisia annua L. Biotechnol Appl Biochem 62(1):17–23. doi:10.1002/bab.1234
Zemach A, McDaniel IE, Silva P, Zilberman D (2010) Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 328:916–919
Zhang YY, Teoh KH, Reed DW, Maes L, Goossens A, Olson DJH, Ross ARS, Covello PS (2008) The molecular cloning of artemisinic aldehyde 11(13) reductase and its role in glandular trichome-dependent biosynthesis of artemisinin in Artemisia annua. J Biol Chem 283:21501–21508
Zhao SS, Zeng MY (1985) Spektrometrische Hochdruck-Flu¨ ssigkeits Chromatographische (HPLC) Untersuchungen zur Analytik von Qinghaosu. Planta Med 51:217–233
Zhao J, Dahle D, Zhou Y, Zhang X, Klibanski A (2005) Hypermethylation of the promoter region is associated with the loss of MEG3 gene expression in human pituitary tumors. J Clin Endocrinol Metab 90(4):2179–2186
Zhu X, Li F, Yang B, Liang J, Qin H, Xu J (2013) Effects of ultraviolet B exposure on DNA methylation in patients with systemic lupus erythematosus. Exp Ther Med 5(4):1219–1225. doi:10.3892/etm.2013.960
Acknowledgments
The authors are thankful to Council of Scientific and Industrial Research (CSIR), New Delhi, India, for financial assistance in the form of project. Authors would like to acknowledge the guidance and support of Dr. Manoj Prasad, National Institute of Plant Genome Research, New Delhi, India, during TLC-analysis. NP is thankful to University Grant Commission (UGC), New Delhi, India for financial assistance in the form of RFSMS (Research fellowship in science for meritorious students).
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
N. Pandey and S. Pandey-Rai have contributed equally.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Pandey, N., Pandey-Rai, S. Deciphering UV-B-induced variation in DNA methylation pattern and its influence on regulation of DBR2 expression in Artemisia annua L.. Planta 242, 869–879 (2015). https://doi.org/10.1007/s00425-015-2323-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-015-2323-3