Abstract
Floral and vegetative development of plants is dependent on the combinatorial action of MADS-domain transcription factors. Members of the STMADS11 subclade, such as MPF1 of Physalis, are abundantly expressed in leaves as well as in floral organs, but their function is not yet clear. Our studies with transgenic Arabidopsis that over-express MPF1 suggest that MPF1 interacts with SOC1 to determine flowering time. However, MPF1 RNAi-mediated knockdown Physalis plants revealed a complex phenotype with changes in flowering time, plant architecture and seed size. Flowering of these plants was delayed by about 20% as compared to wild type. Expression of PFLFY is upregulated in the MPF1 RNAi lines, while PFFT and MPF3 genes are strongly repressed. MPF1 interacts with a subset of MADS-domain factors, namely with PFSOC1 in planta, and with PFSEP3 and PFFUL in yeast, supporting a regulatory role for this protein in flowering. The average size of seeds produced by the transgenic MPF1 RNAi plants is increased almost twofold. The height of these plants is also increased about twofold, but most axillary buds are stunted when compared to controls. Taken together, this suggests that members of the STMADS11 subclade act as positive regulators of flowering but have diverse functions in plant growth.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
MADS-domain proteins play important roles in development and evolution of flowering plants, and often exert their functions as heterooligomers. Engaged both in the control of flowering time and the formation of flowers, they also act outside the flower—for instance, in the control of fruiting and in vegetative development (Theissen 2001; Becker and Theissen 2003). Members of a superclade encompassing four subclades, named STMADS11, STMADS16, SVP and ZMM19, are expressed mostly in vegetative tissues (He and Saedler 2005).
Five genes of the SVP subclade have been functionally characterized so far in dicotyledonous plants: SVP (SHORT VEGETATIVE PHASE) controls flowering time in Arabidopsis thaliana (Hartmann et al. 2000; Lee et al. 2007); JOINTLESS is involved in abscission layer formation (Mao et al. 2000) and suppresses sympodial inflorescence meristem in Solanum lycopersicum (Szymkowiak and Irish 2006); INCO defines a morphological novelty—the prophyll in Antirrhinum majus (Masiero et al. 2004); and PkMADS1 regulates adventitious shoot induction and vegetative shoot development in Paulownia kawakamii (Prakash and Kumar 2002). An SVP ortholog from Eucalyptus grandis, when ectopically expressed in A. thaliana, results in late flowering, suggesting that it may assist in controlling flowering time in its native host as well (Brill and Watson 2004).
Four members of the STMADS16 subclade have been functionally analyzed. Over-expression of STMADS16 from Solanum tuberosum, the founding member of this subclade, affected flowering time and altered inflorescence architecture in transgenic Nicotiana tabacum (Garcia-Maroto et al. 2000). MPF2 controls a morphological novelty, the inflated calyx syndrome (ICS) or “Chinese lantern” in Physalis floridana, and is required for male fertility in this species as well (He and Saedler 2005, 2007b). Over-expression of MPF2, like that of STMADS16, alters inflorescence architecture in transgenic S. tuberosum (He and Saedler 2007a). AGL24, when over-expressed in its native host A. thaliana, promotes flowering and functions as a repressor of floral meristem development (Yu et al. 2002, 2004; Michaels et al. 2003; He and Saedler 2007b). IbMADS4 from sweet potato regulates vegetative shoot development in transgenic chrysanthemum (Aswath et al. 2004).
The ZMM19 subclade comprises all known STMADS11-like sequences from monocots and plays diverse biological functions as well. Over-expression of ZMM19 occurs in the Tunicate mutant of Zea mays, and is responsible for formation of the large glumes encapsulating the maize kernel that give this mutant its name (Münster et al. 2002; He et al. 2004). The phenotype seen in OsMADS22 over-expressing Oryza sativa is reminiscent of Tunicate (Sentoku et al. 2005). In transgenic Arabidopsis, OsMADS22 and its close homolog OsMADS47 cause floral reversions (Fornara et al. 2008). Besides these genes, another homolog, OsMADS55, is also demonstrated to be a negative regulator of brassinosteroid responses (Fornara et al. 2008; Lee et al. 2008b). TaVRT-2 from Triticum aestivum has been shown to code for the repressor of flowering (Kane et al. 2005). Its homologs from barley are demonstrated to inhibit floral meristem identity (Trevaskis et al. 2007).
A common role for these STMADS11 superclade MADS-box genes is in the control of flowering time. Phase transition is a very important developmental process in plants. In Arabidopsis, several key regulators of flowering time, such as CO, FLC and AP1, and key integrators of different floral pathways, including LFY, SOC1 and FT, were functionally characterized and were shown to operate in complicated floral pathways involving transcription regulation and protein–protein interactions (Mandel and Yanofsky 1995; Michaels and Amasino 1999; Wagner et al. 1999; Borner et al. 2000; Lee et al. 2000; Reeves and Coupland 2001; Moon et al. 2003; Parcy 2005; Corbesier et al. 2007).
STMADS11 from S. tuberosum (Carmona et al. 1998) is the founding member of the fourth subclade, which does not include any Arabidopsis proteins (He and Saedler 2005). No function was thus far assigned to members of this subclade. Here, we report molecular and functional characterization of MPF1, a typical member of the STMADS11 subclade, from P. floridana. We provide evidence that MPF1 is involved in plant architecture, seed size and flowering-time control.
Materials and methods
Plant materials
The sources of all solanaceous plant materials involved in this study (S. tuberosum L. and P. floridana Rydb.) are described in He and Saedler (2005). These plants and A. thaliana ecotype Columbia were grown in glasshouses under long- or short-day conditions as indicated.
Isolation of nucleic acids
Leaves and floral organs were harvested and immediately plunged into liquid nitrogen. Genomic DNA of solanaceous plants was isolated with the Dieca buffer (Merck, Darmstadt, Germany). Total RNA was isolated from various organs of both solanaceous plants and Arabidopsis using the total RNA reagent kit (Biomol, Hamburg, Germany).
Isolation of DNA fragments for sequence analysis
Full-length cDNAs, derived from genes of the STMADS11 subclade from various solanaceous species, and partial cDNA sequences for flowering-time genes (except PFSOC1) were isolated using the 5′/3′ RACE Kit (Roche Diagnostics, Mannheim, Germany). Gene-specific primers for members of the STMADS11 subclade were designed based on the STMADS11 cDNA sequence from potato (Carmona et al. 1998). The primers for Physalis flowering-time genes were based on sequences of their homologs from various plant species, obtained from the NCBI database (http://www.ncbi.nlm.nih.gov/). The High Fidelity PCR System (Roche) was used to amplify the cDNAs. Gene-specific primers were designed based on sequences of the full-length cDNAs, and employed to isolate the genomic sequences of the corresponding members of the STMADS11 subclade using the Expand Long Template PCR System (Roche). The promoter sequences were isolated by rapid amplification of gDNA ends analysis using the same primers as for 5′RACE. PCR products were first visualized on 1.0% agarose gels and then purified, or purified directly, with the Highly Pure PCR Product Purification kit (Roche), and cloned into the pGEM T-easy vector (Promega, Madison, WI, USA). Plasmids were prepared using the Miniprep Plasmid Purification kit (Qiagen, Hilden, Germany). Yeast clones were directly characterized using a combination of PCR purification and direct sequencing analysis.
Northern blots
Northern blots were performed as described previously (He et al. 2002). After strip** of probes, the blots were re-hybridized with an ACTIN gene probe from S. tuberosum (He and Saedler 2005). The filters were exposed in a Molecular Dynamics Storage Phosphor Screen and the signals were read with a Typhoon 8600 PhosphorImager (Amersham, Pharmacia Biotech Limited, Little Chalfont, UK).
RT-PCR analysis
Total RNA was isolated from Arabidopsis seedlings of wild-type, MPF1 transgenic and soc1-3 mutant lines, grown under long-day condition. Seedlings were harvested 10, 12 and 14 days after germination. First-strand cDNA was synthesized using reverse transcriptase (Invitrogen, Carlsbad, CA, USA) and PCRs were performed to amplify transcripts derived from the flowering-time control genes CO, FT, SOC1, AP1, LFY, and FLC. For semi-quantitative RT-PCR, the PCR products were run on a 1.0% agarose gel. Real-time RT-PCR was carried out using the Bio-Rad iQ5 Real Time PCR Detection System. Reactions were carried out in 25-μL reactions including 125 nmol/L of gene-specific primer and 1× iQ SYBR Green Supermix solution (Bio-Rad Laboratories, Hercules, CA, USA). The program was set at 95°C for 3 min to activate polymerase, followed by 40 cycles of 95°C for 30 s, 60°C for 30 s, 72°C for 1 min, and a final melting curve analysis from 60 to 95°C. Amplification efficiency was determined and only primer pairs that showed 85–105% amplification efficiency were used for expression studies. The RT-PCR experiments were repeated three times using cDNAs from independent isolations of total RNA (each RT-PCR experiment was repeated three times using the same cDNA). A typical experiment is presented. Relative expression of the flowering-time genes was normalized with respect to PP2A expression according to the Pfaffl method (Bio-Rad).
Transformation of A. thaliana and P. floridana
For over-expression, full-length (sense) MPF1 cDNA was fused to the cauliflower mosaic virus (CaMV) 35S promoter and subsequently cloned into the binary plant vector pBAR-A. For functional promoter analysis, the upstream sequences (from ATG) of STMADS11 (803 and 1,383 bp) and MPF1 (905 and 1,550 bp) were inserted into pGPTV-BAR vector fused with GUS reporter gene. Transformation of Arabidopsis was performed according to Clough and Bent (1998) and transgenic plants were selected by spraying with 0.15% BASTA. Design of constructs, and details of transformations and screening conditions for 35S:MPF1-RNAi P. floridana plants were as described previously (He and Saedler 2005).
GUS histochemical assays of transgenic Arabidopsis
For GUS activity analysis, tissues of GUS reporter transgenic lines were immersed in a staining buffer (100 mM phosphate buffer, 10 mM EDTA, 0.5 mM K3Fe(CN)6, 0.5 mM K4Fe(CN)6, 0.1% Triton X-100 and X-Gluc) and incubated at 37°C overnight. The samples were treated with 70% ethanol repeatedly to remove chlorophyll. The images were taken with a SMZ1000 stereomicroscope (Nikon, Japan).
Characterization and quantification of phenotypes
Bolting time (days after germination), and the numbers of rosette leaves at bolting and anthesis were recorded for wild-type, soc1 and transgenic Arabidopsis lines grown under either short- or long-day conditions. The wild-type and transgenic Physalis were grown under long-day conditions (at 16–22°C), and the overall height and the number of internodes between the cotyledon and the first flower bud, and the length and internode number of each lateral branch were measured at the time of anthesis. At least four plants from each line were included in all cases. Seed weight (g/100 seeds) was balanced three times independently for each line.
Genoty** of transgenic plants
Transgenic Arabidopsis lines were genotyped by Southern blotting and real-time RT-PCR. For real-time RT-PCR, 28-day-old seedlings of wild-type and transgenic Physalis were used. PFMAGO1 (He et al. 2007) was used as an internal control. The procedure and the reaction parameters for RT-PCR analysis were as described for transgenic Arabidopsis.
Yeast two-hybrid assays and library screens
Full-length MPF1 cDNA was cloned into pGBKT7 (Clontech, Laboratories, Mountain View, CA, USA). Transformed yeast cells did not grow on SD/-Trp-His plates, indicating that MPF1 did not self-activate. Full-length MPF1 was, therefore, used as a bait to screen cDNA libraries of Arabidopsis and Physalis (Ling et al. 1995; He et al. 2007). All confirmations were performed on medium-stringency plates containing SD/-Trp-His-Leu and high-stringency plates containing SD/-Trp-His-Leu-Ade, plus 3.0 mM 3-AT, respectively (He et al. 2007). Media were prepared and yeast cells were handled according to standard procedures detailed in the manuals supplied by Clontech. The non-lethal β-galactosidase assay was performed as described by Duttweiler (1996).
Bimolecular fluorescence complementation (BiFC)
Coding regions of MPF1 and PFSOC1 were amplified by PCR with primers that introduced NcoI and EcoRV sites to facilitate the construction of N-terminal fusions in-frame with the N-terminal (aa 1–155) and C-terminal (aa 156–239) domains of yellow fluorescent protein (YFP) in pvk-ctYN and pvk-ctYC, respectively. Four constructs were assembled: pvk-MPF1-ctYC, pvk-MPF1-ctYN, pvk-PFSOC1-ctYC and pvk-PFSOC1-ctYN. The YFP-split pvk vectors (A.Y., unpublished work) essentially correspond to the 3′ module Agroinfection vectors described by Gleba et al. (2004) and Marillonnet et al. (2004). They are able to recombine in plant cells with a viral 5′-module (pICH17388) in the presence of a recombinase vector (pICH10881); these components are provided by separate Agrobacterium tumefaciens GV3101 strains. For the BiFC assay, four Agrobacterium strains were combined: two YFP-split strains (gene fusions in pvk-ctYN and pvk-ctYC) and the 5′ module and recombinase strains. One pvk strain was omitted in negative controls. Seedlings (6–9 weeks old) of P. floridana and Nicotiana benthamiana were used as hosts. Agroinfection was performed as described (Marillonnet et al. 2004), and the fluorescent signal emanating from reconstituted YFP was observed 2–5 days post-infection using a fluorescent microscope (Zeiss Axiophot). Negative controls were also examined with CFP and RFP filter sets.
Results
Heterologous expression couples MPF1 to flowering
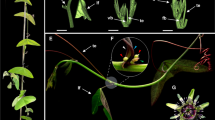
Over-expression of MPF1 in A. thaliana leads to late flowering under both short- and long-day conditions and these 35S:MPF1 transgenic plants seem to phenocopy SOC1 deficiency (Fig. 1a–c; Table 1). Strong and specific interactions of MPF1 with SOC1 were detected in yeast (Fig. 1d), corroborated by the result that only six independent clones of MADS-domain protein SOC1 were uniquely fished out from an Arabidopsis expression library using full-length MPF1 as bait. MPF1 interferes with the expression of flowering-time genes in Arabidopsis. Our data suggest that CO, FLC and AP1 might be primary targets of MPF1 in Arabidopsis under long-day conditions (supplemental Fig. S1). The data from soc1-3 mutants suggest that SOC1 induces the expression of FLC and AP1 at particular times during development (supplemental Fig. S1) and thus seems to share some targets with MPF1. Taking together, these imply that MPF1, directly or indirectly, antagonizes SOC1 function in Arabidopsis and suggest that MPF1 may control flowering time in Physalis as well.
Analysis of over-expressing MPF1 transgenic Arabidopsis plants. a, b Phenotype of a typical 35S:MPF1 transgenic line in comparison to wild-type Arabidopsis grown under long-day (LD) and short-day (SD) conditions, respectively. c RT-PCR analysis of MPF1 gene expression in transgenic Arabidopsis. The upper graph shows data from real-time RT-PCR. The PP2A gene was used as a loading control and for normalization. Error bars represent ±SD (n = 3). The lower panel shows semi-quantitative RT-PCR analysis. d Non-lethal β-galactosidase assays of possible interactions of STMADS11, MSM1 and MPF1 with several other MADS-box proteins (listed on top). The empty vector pGBKT7 and AP2 from Arabidopsis were used as controls
Therefore, expression patterns of MPF1 in Physalis and protein–protein interactions were studied. However, the function of MPF1 most directly is revealed through RNAi MPF1 knockdown experiments.
Expression pattern of the MPF1 gene
We performed Northern blot analysis using gene-specific cDNA fragments as probes. Figure 2a shows that MPF1 mRNA of P. floridana is expressed in all floral whorls, though at different levels, suggesting either that these mRNAs have different turnover rates or that the genes are transcribed at a different level in these organs. In S. tuberosum, which was used as an independent control species, the ortholog STMADS11 was differentially expressed in all floral organs and its mRNA levels were higher in all bud stages than at other stages of flowering (Fig. 2a). In addition, two bands were detected in some floral organs (Fig. 2a), suggesting developmental and post-transcriptional regulation.
MPF1-like gene expression. a Analysis of STMADS11 and MPF1 expression by Northern blotting. ACTIN was used as a loading control. Total RNAs were extracted from leaves (L), sepals, petals, stamens and carpels from buds (B), small buds (sB), large buds (lB), flowers (F), young flowers (yF) or mature flowers (mF), and young fruits (yFr), large fruits (lFr) and old fruits (oFr), respectively. b Functional promoter analysis: GUS expression pattern in STMADS11:GUS (upper) and MPF1:GUS (lower) transgenic Arabidopsis lines. GUS signals are in leaf, inflorescence (flower buds), mature flower and silique, but not in ovary and seeds (from left to right)
We also studied heterologous expression driven by the MPF1 and STMADS11 promoters in Arabidopsis. Two fragments of each promoter (803 and 1,383 bp for STMADS11, and 905 and 1,550 bp for MPF1 that included 5′UTRs) were tested for ability to drive expression of the GUS reported gene in transgenic plants. In spite of the evolutionary distance, size and divergences of sequences tested, the similar GUS expression domains were observed in all cases (Fig. 2b). Although the precise length of the promoters need to be defined, 0.8–1.5 kb fragments upstream of ATG of both genes seem to be sufficient to drive expression in leaves (mainly in veins), flower buds, and all floral organs, though to lower levels (Fig. 2b). However, while GUS signals were also seen in the receptacle of siliques, they were not detectable in seeds (Fig. 2b). This largely reflects the expression patterns of MPF1 and STMADS11 in their native species (Fig. 2a).
The expression of MPF1 in vegetative and, to a lesser extent, in generative tissues is compatible with its involvement in flowering-time control as suggested by the interaction with SOC1 from Arabidopsis. Therefore, MPF1 interacting partners from Physalis were analyzed.
Interacting partners of MPF1 in Physalis
To find candidate Physalis proteins interacting with MPF1, we screened Physalis cDNA expression libraries (He et al. 2007), using MPF1 as a bait (see “Materials and methods”). Sequencing analysis of 15 clones, which were obtained from library screens, revealed that they represented 3 MADS-box genes: five SOC1-like (PFSOC1), seven SEP3-like (PFSEP3) and three FRUITFUL-like (PFFUL) clones.
The MFP1–PFSOC1 interactions were further characterized in the yeast two-hybrid tests and in planta.
MFP1–PFSOC1 interactions in yeast
Sequencing of the five SOC1-like clones isolated from the Physalis library showed that they belonged to two types, coding for two different SOC1-like proteins in Physalis, termed PFSOC1 and PFSOC2. Interaction of MPF1 with PFSOC1 and PFSOC2 was confirmed using a non-lethal β-galactosidase assay in yeast (Fig. 3a). Four PFSOC1 and one PFSOC2 cDNAs were rescued from these colonies. The longest of the PFSOC1 cDNAs recovered from the library screen comprised 957 bp and included 19 bp of 5′UTR. It had a coding capacity of 219 amino acids. The deduced PFSOC1 protein is slightly longer than its Arabidopsis counterpart SOC1 (215 amino acids) and shares 63% identity with Arabidopsis SOC1 at the amino acid level. All PFSOC1 clones encoded complete IKC domains, which are capable of interacting with MPF1, while one of the PFSOC1 cDNAs encoded amino acids 3–156 and displayed only weak interaction with MPF1 (Fig. 3a). This last finding may indicate that the C-terminus of PFSOC1 is important for MPF1 binding.
Characterization of putative target genes and interacting proteins of MPF1 in Physalis. a Non-lethal β-galactosidase assay for interaction between MPF1, the full-length PFSOC (1–219) and its truncated derivatives (gene and sizes are indicated) in yeast. b, c Bimolecular fluorescence complementation (BiFC) assay after agroinfiltration of MPF1 with PFSOC1 into leaf epidermal cells of P. floridana. Fluorescence (green signal) reveals interaction between MPF1 with PFSOC1 in planta. The relevant combinations of complementation constructs are indicated in the panels. Arrows indicate the nuclei
Only a partial sequence of PFSOC2 was obtained from the library and, therefore, this gene was not studied further, although it seemed to comprise the IKC domain as well.
MPF1 interacts with PFSOC1 in planta
The interaction of MPF1 and PFSOC1 in planta was assessed by BiFC using YFP-split technology (Hu et al. 2002; Hu and Kerppola 2003; Bracha-Drori et al. 2004; Walter et al. 2004; see “Materials and methods”), in which the interaction of two proteins is indicated by the reconstitution of YFP fluorescence. For this purpose, the N- and C-terminal halves of YFP (YN and YC), which exhibit no auto-fluorescence, were fused in-frame to MPF1 and PFSOC1, respectively, and the two fusion proteins were co-expressed in transgenic plants. For co-expression assays, we used a viral expression system based on in planta engineering of viral RNA replicons (Gleba et al. 2004; Marillonnet et al. 2004). Using Agroinfection, N- or C-terminal fusions with the split YFP fragments were transiently co-expressed in leaves of P. floridana and N. benthamiana. Co-expression of either YN-MPF1/YC-PFSOC1 or YN-PFSOC1/YC-MPF1 yielded a YFP signal in both nucleus and cytoplasm (Fig. 3b, c). No such signal was observed in controls. Thus, these data suggest that MPF1 and PFSOC1 form heterodimers in planta and confirm the results of the yeast two-hybrid assays. In addition, these results further strengthen the involvement of MPF1 in flowering-time control.
However, the function of a gene can be best inferred from its mutant phenotype. Unfortunately, no such mutants are available in P. floridana; therefore, we decided to use RNAi technology to silence the MPF1 gene.
Analysis of 35S:MPF1-RNAi transgenic P. floridana plants
We obtained 12 35S:MPF1 RNAi transgenic P. floridana lines and genotyped them by real-time RT-PCR (Fig. 4a). The morphology of the knockdown (KO) lines that displayed the greatest degree of suppression of MPF1 (i.e. KO2, KO3, KO5, KO6, KO9 and KO10) was further examined.
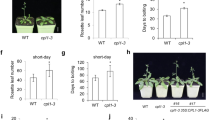
Expression of MPF1 and flowering time of 35S:MPF1-RNAi transgenic Physalis lines. a Expression of MPF1 in 35S:MPF1-RNAi lines. MPF1 expression was measured in the 12 knockdown (KO) transgenic lines using real-time RT-PCR, and expression levels were normalized relative to wild-type (WT). Error bars represent ±SD (n = 3). b Flowering time of the transgenic 35S:MPF1-RNAi lines and wild-type Physalis. Flowering time is given in days to anthesis. The black column represents wild type and the gray columns represent individual transgenic lines. Days 0–30 were condensed to allow magnification of days 30–70 for apparent differences. Error bars represent ±SD (n = 4). c The expression levels of the putative flowering-time genes (from left to right) in wild-type Physalis (black column) and the transgenic knockdown lines KO2 (dark gray), KO3 (middle gray), KO4 (light gray) and KO10 (white). Plant lines were grown for 28 days under long-day conditions. The expression level of each gene was normalized relative to wild type (black column). Error bars represent ±SD (n = 3)
Flowering time and expression of putative flowering-time genes
No floral abnormalities were observed but most of these KO lines flowered on average 10 days later than the wild type (Fig. 4b). To verify that MPF1 plays a positive role in determining flowering time, we examined expression of putative Physalis orthologs of various Arabidopsis genes that are known to participate in flowering.
We isolated from Physalis PFCO (CO-like), PFFT (FT-like), PFSOC1 (SOC1-like), PFLFY (LFY-like) and MPF3 (AP1-like). All attempts to isolate an FLC-like sequence failed, implying that a close homolog does not exist in Physalis. The expression profiles of the above genes were investigated by real-time RT-PCR in 28-day-old seedlings from wild-type Physalis and from the four MPF1 RNAi lines KO2, KO3, KO4 and KO10. KO2, KO3 and KO10 showed severe flowering phenotype (Fig. 4a), whereas KO4 manifested no obvious phenotype (Fig. 4a) and expressed MPF1 at a wild-type level. In accordance with these flowering phenotypes, the PFLFY gene was significantly upregulated, while PFFT and MPF3 were strongly repressed in KO2, KO3 and KO10, compared to wild type and KO4 (Fig. 4c).
Plant architecture
As shown in Fig. 5a, the overall height and the distance between internodes were dramatically increased in the six transgenic plants with the most severe phenotype (by up to 82 and 74%, respectively). However, the number of internodes on the main stem was unchanged (Fig. 5a, b). In wild-type Physalis, the axillary shoots were fully developed before flowering, while in the MPF1 knockdown lines (KO2, KO3, KO5, KO6, KO9 and KO10), most axillary buds were stunted (Fig. 5a, b). Furthermore, this seemed to depend on the position of the shoot on the main stem (Fig. 5a, b). The cotyledonous axillary shoots were markedly reduced in size, but the first two axillary lateral shoots remained unaffected as compared to wild type. Internode numbers on later-emerging lateral shoots were increased, but internodes were closer together than in the wild type, resulting in reduced overall shoot lengths (Fig. 5a, b). Thus, MPF1 seems to act as a repressor for main stem growth and as an activator of lateral shoots.
Plant architecture, seed size and seed weight in transgenic Physalis. a Plant architecture. The most severely affected 35S:MPF1-RNAi lines (KO2, KO3, KO5, KO6, KO9 and KO10) were selected, and their vegetative development was examined when the first flower opened. Average lengths of the elements indicated were determined for each of the six lines and normalized relative to wild type. The data points shown as black dots indicate relative overall lengths. Internode number is given by gray squares and internode length by gray triangles. MS is the main stem, CS1 and CS2 indicate cotyledonous axillary side shoots and LS1 to LS7 are axillary side shoots of the main stem. b Schematic representation of growth habit of 35S:MPF1-RNAi and wild-type plants. DB is first dichotomous branch (other abbreviations as in a); the black dot indicates the first flower. Arrowheads show the direction of growth. c Comparison of seed size in wild type and the KO2 line. d Seed weight in transgenic Physalis lines (Fig. 4a)
Seed size
Mutant seeds of KO2 are significantly bigger than those of wild type (Fig. 5c). This holds for all KO lines in which MPF1 expression is reduced significantly (Fig. 5d), but it also includes line KO11 featuring an intermediate expression level of MPF1 (Fig. 4a). The seed viability seems not to be affected by knockdown of MPF1 since their germination rate is similar to that of wild-type seed (data not shown).
In summary, these results indicate that, in addition to controlling flowering time in Physalis, MPF1 inhibits growth of the main stem, promotes development of axillary shoots and retards seed development.
Discussion
MADS-domain transcription factors act as homo- or heterodimers and often affect more than one trait (Schwarz-Sommer et al. 1990; Coen and Meyerowitz 1991; Weigel and Meyerowitz 1994; Yu et al. 2002, 2004; He and Saedler 2005; Melzer et al. 2008). Here, we demonstrate that MPF1, a member of STMADS11 subclade MADS-box genes, also plays multiple roles in Physalis:
MPF1 acts as a promoter of flowering in Physalis
MPF1 apparently promotes flowering in Physalis. The direct evidence for this comes from experiments that revealed about 20% delay in flowering time in MPF1 RNAi knockdown lines. MPF1 could exert its role in flowering either via a direct regulation of flowering-time control genes or through interaction with their proteins. This assumption is supported by the observation that MPF1 can interact with the products of MADS-box genes, such as PFSOC1, PFFUL and PFSEP3 from Physalis, the orthologs of which are all involved in the control of flowering in Arabidopsis (Borner et al. 2000; Lee et al. 2000; Teper-Bamnolker and Samach 2005), and that MPF1 promoted expression of the putative orthologs of FT (PFFT), AP1 (MPF3) and, to a lesser extent, SOC1 (PFSOC1) and CO (PFCO) in Physalis. A relative loss of activation of these might contribute to the late-flowering phenotype of the MPF1 knockdown mutants. On the other hand, MPF1 repressed PFLFY (the ortholog of LFY); release from this repression should promote flowering and thus might limit the delay in the knockdown mutant to the extent actually observed. In addition, FLC-like genes, repressors of flowering in Arabidopsis (Michaels and Amasino 1999; Ratcliffe et al. 2001, 2003), seem to be missing in the Physalis species.
Remarkably, MPF1 acts as a repressor of flowering and affected many genes such as CO, FLC and AP1 involved in its control either directly or through interaction with SOC1 when over-expressed in the heterologous host, i.e. Arabidopsis. We found that MPF1 induced CO transiently and FLC more stably in transgenic Arabidopsis, and thereby acted to repress FT and SOC1, which could account for much of the delay in flowering time. The repression of AP1 might further enhance the delay in flowering.
This scenario is corroborated by the observations that MPF1 interacts differently with other MADS-domain proteins from Physalis and Arabidopsis. In Physalis, MPF1 interacted with PFSOC1, PFSEP3 and PFFUL. Their Arabidopsis orthologs are known to be involved in flowering (Borner et al. 2000; Lee et al. 2000; Teper-Bamnolker and Samach 2005). However, when expressed in Arabidopsis, MPF1 is found to interact only with SOC1, an integrator of floral pathways (Lee et al. 2000; Moon et al. 2003).
SVP and AGL24 in Arabidopsis are two close homologs of MPF1 but they are not orthologs (He and Saedler 2005) and they are involved in flowering-time control in Arabidopsis (Hartmann et al. 2000; Yu et al. 2002; Michaels et al. 2003; Lee et al. 2007). Both proteins share a similar spectrum of interacting partners such as SEP3, SOC1, FUL, AP1, etc. (Masiero et al. 2004; de Folter et al. 2005) and SVP interacts directly with FLC (Fujiwara et al. 2008) as well. These interactions (i.e. with SOC1) seem to be most important for their integration into floral pathways (Masiero et al. 2004; de Folter et al. 2005; Fujiwara et al. 2008; Lee et al. 2008a; Liu et al. 2008). As a close homolog of SVP and AGL24, MPF1 shares most of their interaction profiles with PFSOC1, PFSEP3 and PFFUL in Physalis but not with corresponding orthologs of the latter from Arabidopsis.
These findings might suggest that the basic structure of the “regulatory and interacting” networks controlling flowering time has been conserved during evolution, but details have diverged substantially between these fairly distant species, A. thaliana of the Brassicaceae and P. floridana of the Solanaceae.
MPF1 controls plant architecture and seed development
MPF1 functions are not restricted to flowering. Transgenic knockdown plants were taller and more slender than wild-type Physalis. While MPF1 seemed to repress growth of the main stem, axillary shoot development was promoted to different degrees along the plant axis. Cotyledonous axillary shoots of MPF1-silenced plants were smaller than those of wild-type Physalis, first and second lateral shoots were normal and the other laterals were reduced in length. Thus, MPF1 seems to play an important role in plant architecture. Recently, Melzer et al. (2008) have shown that the soc1 ful double mutant of Arabidopsis features a very different plant habitus compared to wild type, i.e. exaggerated indeterminate growth. Since, however, Physalis MPF1 interacts only with SOC1 and not with FUL of Arabidopsis, this may explain the lack of a corresponding phenotype when over-expressing the Physalis gene in the heterologous host. However, the situation in Physalis itself is different. Here, MPF1 interacts with both PFSOC1 and PFFUL and hence downregulation of MPF1 might be the reason for generating the architectural plant phenotype observed. Since too little genomics has been done in Physalis thus far and no microarrays are yet available, the components controlling plant architecture remain elusive.
The third trait controlled by MPF1 in Physalis is seed development. Seed weight and size were doubled in lines in which MPF1 was severely downregulated, suggesting that MPF1 acts as a repressor. The contribution of about seven different MADS-domain proteins to fruit and seed development has been studied in S. lycopersicum (Busi et al. 2003), a relative of Physalis. These authors suggested that TAGL2 could represent a SEPALLATA function in tomato and, through an interacting network with other MADS-domain proteins, especially TAG1, might control seed development. Since we showed that MPF1 interacts with PFSEP3, it can probably affect seed development in Physalis in a similar fashion. However, the interaction of MPF1 with PFFUL, an ortholog of FUL, might provide an alternative. Seeds of Arabidopsis ful mutants are slightly smaller than wild type (Gu et al. 1998), while 35S:FUL Arabidopsis plants not only flower early, but also feature a threefold higher seed weight when compared to wild type (Ferrandiz et al. 2000). It is conceivable, therefore, that increased seed size and seed weight in the MPF1-RNAi knockdown Physalis plants might reflect reduced levels of the MPF1/PFFUL complex and relative increase of PFFUL, compared to wild type. The seed/fruit phenotype of transgenic Physalis over-expressing PFFUL might substantiate this speculative scenario.
In conclusion, we functionally characterized MPF1 in Physalis, a member of the STMADS11 subclade encoding hypothetical MADS-domain transcriptional factors. Our data validate that MPF1 controls flowering time, plant architecture and seed development in Physalis. Our work additionally suggests that caution has to be taken when the function of a protein involved in a particular network in its native host is studied in an evolutionary distantly related model plant.
Abbreviations
- RNAi:
-
RNA interference
- BiFC:
-
Bimolecular fluorescence complementation
- YFP:
-
Yellow fluorescent protein
- KO:
-
Knockdown
References
Aswath CR, Mo SY, Kim SH, Kim DH (2004) IbMADS4 regulates the vegetative shoot development in transgenic chrysanthemum (Dendrathema grandiflora (Ramat.) Kitamura). Plant Sci 166:847–854
Becker A, Theissen G (2003) The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol Phylogenet Evol 29:464–489
Borner R, Kampmann G, Chandler J, Gleissner R, Wisman E, Apel K, Melzer S (2000) A MADS domain gene involved in the transition to flowering in Arabidopsis. Plant J 24:591–599
Bracha-Drori K, Shichrur K, Katz A, Oliva M, Angelovici R, Yalovsky S, Ohad N (2004) Detection of protein–protein interactions in plants using bimolecular fluorescence complementation. Plant J 40:419–427
Brill EM, Watson JM (2004) Ectopic expression of a Eucalyptus grandis SVP orthologue alters the flowering time of Arabidopsis thaliana. Funct Plant Biol 31:217–224
Busi MV, Bustamante C, D’Angelo C, Hidalgo-Cuevas M, Boggio SB, Valle EM, Zabaleta E (2003) MADS-box genes expressed during tomato seed and fruit development. Plant Mol Biol 52:801–815
Carmona MJ, Ortega N, Garcia-Maroto F (1998) Isolation and molecular characterization of a new vegetative MADS-box gene from Solanum tuberosum L. Planta 207:181–188
Clough SJ, Bent A (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Coen ES, Meyerowitz EM (1991) The war of the whorls: genetic interactions controlling flower development. Nature 353:31–37
Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, Coupland G (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316:1030–1033
de Folter S, Immink RG, Kieffer M, Parenicová L, Henz SR, Weigel D, Busscher M, Kooiker M, Colombo L, Kater MM, Davies B, Angenent GC (2005) Comprehensive interaction map of the Arabidopsis MADS box transcription factors. Plant Cell 17:1424–1433
Duttweiler HM (1996) A highly sensitive and non-lethal β-galactosidase plate assay for yeast. Trends Genet 12:340–341
Ferrandiz C, Liljegren SJ, Yanofsky MF (2000) Negative regulation of the SHATTERPROOF genes by FRUITFULL during Arabidopsis fruit development. Science 289:436–438
Fornara F, Gregis V, Pelucchi N, Colombo L, Kater M (2008) The rice StMADS11-like genes OsMADS22 and OsMADS47 cause floral reversions in Arabidopsis without complementing the svp and agl24 mutants. J Exp Bot 59:2181–2190
Fujiwara S, Oda A, Yoshida R, Niinuma K, Miyata K, Tomozoe Y, Tajima T, Nakagawa M, Hayashi K, Coupland G, Mizoguchi T (2008) Circadian clock proteins LHY and CCA1 regulate SVP protein accumulation to control flowering in Arabidopsis. Plant Cell 20:2960–2971
Garcia-Maroto F, Ortega N, Lozano R, Carmona MJ (2000) Characterization of the potato MADS-box gene STMADS16 and expression analysis in tobacco transgenic plants. Plant Mol Biol 42:499–513
Gleba Y, Marillonnet S, Klimyuk V (2004) Engineering viral expression vectors for plants: the ‘full virus’ and the ‘deconstructed virus’ strategies. Curr Opin Plant Biol 7:182–188
Gu Q, Ferrándiz C, Yanofsky MF, Martienssen R (1998) The FRUITFULL MADS-box gene mediates cell differentiation during Arabidopsis fruit development. Development 125:1509–1517
Hartmann U, Hohmann S, Nettesheim K, Wisman E, Saedler H, Huijser P (2000) Molecular cloning of SVP: a negative regulator of the floral transition in Arabidopsis. Plant J 21:351–360
He CY, Saedler H (2005) Heterotopic expression of MPF2 is the key to the evolution of the Chinese lantern of Physalis, a morphological novelty in Solanaceae. Proc Natl Acad Sci USA 102:5779–5784
He CY, Saedler H (2007a) Molecular evolution of a morphological novelty in Solanaceae, the Inflated-Calyx-Syndrome (ICS) in Physalis. Acta Hort 745:171–182
He CY, Saedler H (2007b) Hormonal control of the inflated calyx syndrome, a morphological novelty, in Physalis. Plant J 49:935–946
He CY, Zhang J, Chen S (2002) A soybean gene encoding a proline-rich protein is regulated by salicylic acid, an endogenous circadian rhythm and by various stresses. Theor Appl Genet 104:1125–1131
He CY, Münster T, Saedler H (2004) On the origin of floral morphological novelties. FEBS Lett 567:147–151
He CY, Sommer H, Grosardt B, Huijser P, Saedler H (2007) PFMAGO, a MAGO NASHI-like factor, interacts with the MADS-box protein MPF2 from Physalis floridana. Mol Biol Evol 24:1229–1241
Hu CD, Kerppola TK (2003) Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat Biotechnol 21:539–545
Hu CD, Chinenov Y, Kerppola TK (2002) Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell 9:789–798
Kane NA, Danyluk J, Tardif G, Ouellet F, Laliberte JF, Limin AE, Fowler DB, Sarhan F (2005) TaVRT-2, a member of the StMADS-11 clade of flowering repressors, is regulated by vernalization and photoperiod in wheat. Plant Physiol 138:2354–2363
Lee H, Suh SS, Park E, Cho E, Ahn JH, Kim SG, Lee JS, Kwon YM, Lee I (2000) The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev 14:2366–2376
Lee JH, Yoo SJ, Park SH, Hwang I, Lee JS, Ahn JH (2007) Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev 21:397–402
Lee J, Oh M, Park H, Lee I (2008a) SOC1 translocated to the nucleus by interaction with AGL24 directly regulates leafy. Plant J 55:832–843
Lee S, Choi SC, An G (2008b) Rice SVP-group MADS-box proteins, OsMADS22 and OsMADS55, are negative regulators of brassinosteroid response. Plant J 54:93–105
Ling M, Merante F, Robison BR (1995) A rapid and reliable DNA preparation method for screening a large number of yeast clones by polymerase chain reaction. Nucleic Acids Res 23:4924–4925
Liu C, Chen H, Er HL, Soo HM, Kumar PP, Han JH, Liou YC, Yu H (2008) Direct interaction of AGL24 and SOC1 integrates flowering signals in Arabidopsis. Development 135:1481–1491
Mandel MA, Yanofsky MF (1995) A gene triggering flower formation in Arabidopsis. Nature 377:522–524
Mao L, Begum D, Chuang H, Budiman MA, Szymkowiak EJ, Irish EE, Wing RA (2000) JOINTLESS is a MADS-box gene controlling tomato flower abscission zone development. Nature 406:910–913
Marillonnet S, Giritch A, Gils M, Kandzia R, Klimyuk V, Gleba Y (2004) In planta engineering of viral RNA replicons: efficient assembly by recombination of DNA modules delivered by Agrobacterium. Proc Natl Acad Sci USA 101:6852–6857
Masiero S, Li MA, Will I, Hartmann U, Saedler H, Huijser P, Schwarz-Sommer Zs, Sommer H (2004) INCOMPOSITA: a MADS-box gene controlling prophyll development and floral meristem identity in Antirrhinum. Development 131:5981–5990
Melzer S, Lens F, Gennen J, Vanneste S, Rohde A, Beeckman T (2008) Flowering-time genes modulate meristem determinacy and growth form in Arabidopsis thaliana. Nat Genet 40:1489–1492
Michaels SD, Amasino RM (1999) FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11:949–956
Michaels SD, Ditta G, Gustafson-Brown C, Pelaz S, Yanofsky M, Amasino RM (2003) AGL24 acts as a promoter of flowering in Arabidopsis and is positively regulated by vernalization. Plant J 33:867–874
Moon J, Suh SS, Lee H, Choi KR, Hong CB, Paek NC, Kim SG, Lee I (2003) The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J 35:613–623
Münster T, Deleu W, Wingen LU, Ouzunova M, Cacharrón J, Faigl W, Werth S, Kim JTT, Saedler H, Theiβen G (2002) Maize MADS-box genes galore. Maydica 47:287–301
Parcy F (2005) Flowering: a time for integration. Int J Dev Biol 49:585–593
Prakash AP, Kumar PP (2002) PkMADS1 is a novel MADS box gene regulating adventitious shoot induction and vegetative shoot development in Paulownia kawakamii. Plant J 29:141–151
Ratcliffe OJ, Nadzan GC, Reuber TL, Riechmann JL (2001) Regulation of flowering in Arabidopsis by an FLC homologue. Plant Physiol 126:122–132
Ratcliffe OJ, Kumimoto RW, Wong BJ, Riechmann JL (2003) Analysis of the Arabidopsis MADS AFFECTING FLOWERING gene family: MAF2 prevents vernalization by short periods of cold. Plant Cell 15:1159–1169
Reeves PH, Coupland G (2001) Analysis of flowering time control in Arabidopsis by comparison of double and triple mutants. Plant Physiol 126:1085–1091
Schwarz-Sommer Z, Huijser P, Nacken W, Saedler H, Sommer H (1990) Genetic control of flower development by homeotic genes in Antirrhinum majus. Science 250:931–936
Sentoku N, Kato H, Kitano H, Imai R (2005) OsMADS22, an STMADS11-like MADS-box gene of rice, is expressed in non-vegetative tissues and its ectopic expression induces spikelet meristem indeterminacy. Mol Genet Genom 273:1–9
Szymkowiak EJ, Irish EE (2006) JOINTLESS suppresses sympodial identity in inflorescence meristems of tomato. Planta 223:646–658
Teper-Bamnolker P, Samach A (2005) The flowering integrator FT regulates SEPALLATA3 and FRUITFULL accumulation in Arabidopsis leaves. Plant Cell 17:2661–2675
Theissen G (2001) Development of floral organ identity: stories from the MADS house. Curr Opin Plant Biol 4:75–85
Trevaskis B, Tadege M, Hemming MN, Peacock WJ, Dennis ES, Sheldon C (2007) SVP-like MADS-box genes inhibit floral meristem identity in barley. Plant Physiol 143:225–235
Wagner D, Sablowski RW, Meyerowitz EM (1999) Transcriptional activation of APETALA1 by LEAFY. Science 285:582–584
Walter M, Chaban C, Schutze K, Batistic O, Weckermann K, Nake C, Blazevic D, Grefen C, Schumacher K, Oecking C, Harter K, Kudla J (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40:428–438
Weigel D, Meyerowitz EM (1994) The ABCs of floral homeotic genes. Cell 78:203–209
Yu H, Xu Y, Tan EL, Kumar PP (2002) AGAMOUS-like 24, a dosage-dependent mediator of the flowering signals. Proc Natl Acad Sci USA 99:16336–16341
Yu H, Ito T, Wellmer F, Meyerowitz EM (2004) Repression of AGAMOUS-LIKE 24 is a crucial step in promoting flower development. Nat Genet 36:157–161
Acknowledgments
We thank Dr. Peter Huijser (Max-Planck Institute for Plant Breeding Research, Cologne, Germany) for providing soc1-3 mutant seeds and for his constructive suggestions and Dr. Zheng Meng (Institute of Botany, Chinese Academy of Sciences, Bei**g, China) for his valuable comments to improve the manuscript. We also thank Mrs. Susanne Hoehmann and Nora Bujdoso (Max-Planck Institute for Plant Breeding Research, Cologne, Germany) for sharing some Arabidopsis primers for real-time RT-PCRs. The technical assistance from Britta Grosardt (Max-Planck Institute for Plant Breeding Research, Cologne, Germany) and Pichang Gong (Institute of Botany, Chinese Academy of Sciences, Bei**g, China) is also acknowledged. This work was partially supported by the Hundred Talents Project of the CAS to CYH.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
He, C., Tian, Y., Saedler, R. et al. The MADS-domain protein MPF1 of Physalis floridana controls plant architecture, seed development and flowering time. Planta 231, 767–777 (2010). https://doi.org/10.1007/s00425-009-1087-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-009-1087-z