Abstract
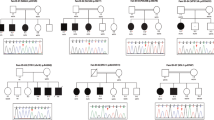
Mutations or structural genomic alterations of the X-chromosomal gene ARHGEF9 have been described in male and female patients with intellectual disability. Hyperekplexia and epilepsy were observed to a variable degree, but incompletely described. Here, we expand the phenotypic spectrum of ARHGEF9 by describing a large Ethiopian-Jewish family with epilepsy and intellectual disability. The four affected male siblings, their unaffected parents and two unaffected female siblings were recruited and phenotyped. Parametric linkage analysis was performed using SNP microarrays. Variants from exome sequencing in two affected individuals were confirmed by Sanger sequencing. All affected male siblings had febrile seizures from age 2–3 years and intellectual disability. Three developed afebrile seizures between age 7–17 years. Three showed focal seizure semiology. None had hyperekplexia. A novel ARHGEF9 variant (c.967G>A, p.G323R, NM_015185.2) was hemizygous in all affected male siblings and heterozygous in the mother. This family reveals that the phenotypic spectrum of ARHGEF9 is broader than commonly assumed and includes febrile seizures and focal epilepsy with intellectual disability in the absence of hyperekplexia or other clinically distinguishing features. Our findings suggest that pathogenic variants in ARHGEF9 may be more common than previously assumed in patients with intellectual disability and mild epilepsy.

Similar content being viewed by others
References
Körber C, Richter A, Kaiser M et al (2012) Effects of distinct collybistin isoforms on the formation of GABAergic synapses in hippocampal neurons. Mol Cell Neurosci 50:250–259. doi:10.1016/j.mcn.2012.05.006
Machado Camila Oliveira, Freitas Griesi-Oliveira K, Rosenberg C et al (2016) Collybistin binds and inhibits mTORC1 signaling: a potential novel mechanism contributing to intellectual disability and autism. Eur J Hum Genet 24:59–65. doi:10.1038/ejhg.2015.69
Harvey K, Duguid IC, Alldred MJ et al (2004) The GDP-GTP exchange factor collybistin: an essential determinant of neuronal gephyrin clustering. J Neurosci 24:5816–5826. doi:10.1523/JNEUROSCI.1184-04.2004
Marco EJ, Abidi FE, Bristow J et al (2008) ARHGEF9 disruption in a female patient is associated with X linked mental retardation and sensory hyperarousal. J Med Genet 45:100–105. doi:10.1136/jmg.2007.052324
Shimojima K, Sugawara M, Shichiji M et al (2011) Loss-of-function mutation of collybistin is responsible for X-linked mental retardation associated with epilepsy. J Hum Genet 56:561–565. doi:10.1038/jhg.2011.58
Lemke JR, Riesch E, Scheurenbrand T et al (2012) Targeted next generation sequencing as a diagnostic tool in epileptic disorders. Epilepsia 53:1387–1398. doi:10.1111/j.1528-1167.2012.03516.x
de Ligt J, Willemsen MH, van Bon Bregje W M et al (2012) Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med 367:1921–1929. doi:10.1056/NEJMoa1206524
Long P, May MM, James VM et al (2015) Missense mutation R338W in ARHGEF9 in a family with X-linked intellectual disability with variable macrocephaly and macro-orchidism. Front Mol Neurosci 8:83. doi:10.3389/fnmol.2015.00083
Johnson JP, Nelson R, Schwartz CE (1998) A family with mental retardation, variable macrocephaly and macro-orchidism, and linkage to Xq12-q21. J Med Genet 35:1026–1030
Lesca G, Till M, Labalme A et al (2011) De novo Xq11.11 microdeletion including ARHGEF9 in a boy with mental retardation, epilepsy, macrosomia, and dysmorphic features. Am J Med Genet A 155A:1706–1711. doi:10.1002/ajmg.a.34004
Kalscheuer VM, Musante L, Fang C et al (2009) A balanced chromosomal translocation disrupting ARHGEF9 is associated with epilepsy, anxiety, aggression, and mental retardation. Hum Mutat 30:61–68. doi:10.1002/humu.20814
Holman SK, Morgan T, Baujat G et al (2013) Osteopathia striata congenita with cranial sclerosis and intellectual disability due to contiguous gene deletions involving the WTX locus. Clin Genet 83:251–256. doi:10.1111/j.1399-0004.2012.01905.x
Bhat G, LaGrave D, Millson A et al (2016) Xq11.1-11.2 deletion involving ARHGEF9 in a girl with autism spectrum disorder. Eur J Med. doi:10.1016/j.ejmg.2016.05.014
Purcell S, Neale B, Todd-Brown K et al (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81:559–575. doi:10.1086/519795
Bahlo M, Bromhead CJ (2009) Generating linkage map** files from Affymetrix SNP chip data. Bioinformatics 25:1961–1962. doi:10.1093/bioinformatics/btp313
Abecasis GR, Cherny SS, Cookson WO et al (2002) Merlin–rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30:97–101. doi:10.1038/ng786
Yang H, Wang K (2015) Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat Protoc 10:1556–1566. doi:10.1038/nprot.2015.105
Adzhubei IA, Schmidt S, Peshkin L et al (2010) A method and server for predicting damaging missense mutations. Nat Methods 7:248–249. doi:10.1038/nmeth0410-248
Kumar P, Henikoff S, Ng PC (2009) Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 4:1073–1081. doi:10.1038/nprot.2009.86
Kircher M, Witten DM, Jain P et al (2014) A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 46:310–315. doi:10.1038/ng.2892
The 1000 Genomes Project Consortium (2012) An integrated map of genetic variation from 1,092 human genomes. Nature 491:56–65. doi:10.1038/nature11632
Scheffer IE, Heron SE, Regan BM et al (2014) Mutations in mTOR regulator DEPDC5 cause focal epilepsy with brain malformations. Ann Neurol 75:782–787. doi:10.1002/ana.24126
Baulac S, Ishida S, Marsan E et al (2015) Familial focal epilepsy with focal cortical dysplasia due to DEPDC5 mutations. Ann Neurol 77:675–683. doi:10.1002/ana.24368
Ricos MG, Hodgson BL, Pippucci T et al (2016) Mutations in the mammalian target of rapamycin pathway regulators NPRL2 and NPRL3 cause focal epilepsy. Ann Neurol 79:120–131. doi:10.1002/ana.24547
Sim JC, Scerri T, Fanjul-Fernández M et al (2016) Familial cortical dysplasia caused by mutation in the mammalian target of rapamycin regulator NPRL3. Ann Neurol 79:132–137. doi:10.1002/ana.24502
Weckhuysen S, Marsan E, Lambrecq V et al (2016) Involvement of GATOR complex genes in familial focal epilepsies and focal cortical dysplasia. Epilepsia 57:994–1003. doi:10.1111/epi.13391
Acknowledgements
We thank the family for participation in our study. The study was supported by the Deutsche Forschungsgemeinschaft with a Trilateral Grant (HE 5415/5-1) and within the EuroEPINOMICS project (HE 5415/3-1), the Christian-Albrechts-University Kiel, the Detlev-Wrobel-Fonds for Epilepsy Research Frankfurt, the Klaus-Dieter Scharf-Forschungsprojekt and the Share Value Stiftung.
Author information
Authors and Affiliations
Consortia
Corresponding author
Ethics declarations
Conflicts of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest with respect to this publication.
Ethical standards
The study was approved by the Sheba Medical Center Helsinki Committee and the ethics committee of the University of Kiel and has, therefore, been performed in accordance with the ethical standards laid down in the 1964 Declaration of Helsinki and its later amendments. All participants or their legal guardians provided written informed consent.
Rights and permissions
About this article
Cite this article
Klein, K.M., Pendziwiat, M., Eilam, A. et al. The phenotypic spectrum of ARHGEF9 includes intellectual disability, focal epilepsy and febrile seizures. J Neurol 264, 1421–1425 (2017). https://doi.org/10.1007/s00415-017-8539-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00415-017-8539-3