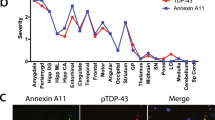
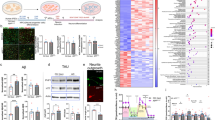
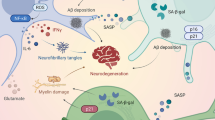
Abstract
Selective neuronal vulnerability to protein aggregation is found in many neurodegenerative diseases including Alzheimer’s disease (AD). Understanding the molecular origins of this selective vulnerability is, therefore, of fundamental importance. Tau protein aggregates have been found in Wolframin (WFS1)-expressing excitatory neurons in the entorhinal cortex, one of the earliest affected regions in AD. The role of WFS1 in Tauopathies and its levels in tau pathology-associated neurodegeneration, however, is largely unknown. Here we report that WFS1 deficiency is associated with increased tau pathology and neurodegeneration, whereas overexpression of WFS1 reduces those changes. We also find that WFS1 interacts with tau protein and controls the susceptibility to tau pathology. Furthermore, chronic ER stress and autophagy-lysosome pathway (ALP)-associated genes are enriched in WFS1-high excitatory neurons in human AD at early Braak stages. The protein levels of ER stress and autophagy-lysosome pathway (ALP)-associated proteins are changed in tau transgenic mice with WFS1 deficiency, while overexpression of WFS1 reverses those changes. This work demonstrates a possible role for WFS1 in the regulation of tau pathology and neurodegeneration via chronic ER stress and the downstream ALP. Our findings provide insights into mechanisms that underpin selective neuronal vulnerability, and for develo** new therapeutics to protect vulnerable neurons in AD.








Similar content being viewed by others
References
Abisambra JF, **wal UK, Blair LJ, O’Leary JC 3rd, Li Q, Brady S et al (2013) Tau accumulation activates the unfolded protein response by impairing endoplasmic reticulum-associated degradation. J Neurosci 33:9498–9507. https://doi.org/10.1523/JNEUROSCI.5397-12.2013
Asai H, Ikezu S, Tsunoda S, Medalla M, Luebke J, Haydar T et al (2015) Depletion of microglia and inhibition of exosome synthesis halt tau propagation. Nat Neurosci 18:1584–1593. https://doi.org/10.1038/nn.4132
Beach TG, Adler CH, Sue LI, Serrano G, Shill HA, Walker DG et al (2015) Arizona study of aging and neurodegenerative disorders and brain and body donation program. Neuropathology 35:354–389. https://doi.org/10.1111/neup.12189
Boland B, Yu WH, Corti O, Mollereau B, Henriques A, Bezard E et al (2018) Promoting the clearance of neurotoxic proteins in neurodegenerative disorders of ageing. Nat Rev Drug Discov 17:660–688. https://doi.org/10.1038/nrd.2018.109
Bolos M, Llorens-Martin M, Jurado-Arjona J, Hernandez F, Rabano A, Avila J (2016) Direct evidence of internalization of tau by microglia in vitro and in vivo. J Alzheimers Dis 50:77–87. https://doi.org/10.3233/JAD-150704
Braak H, Braak E (1991) Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol 82:239–259. https://doi.org/10.1007/BF00308809
Cagalinec M, Liiv M, Hodurova Z, Hickey MA, Vaarmann A, Mandel M et al (2016) Role of mitochondrial dynamics in neuronal development: mechanism for Wolfram syndrome. PLoS Biol 14:e1002511. https://doi.org/10.1371/journal.pbio.1002511
Chen S, Chang Y, Li L, Acosta D, Morrison C, Wang C et al (2021) Spatially resolved transcriptomics reveals unique gene signatures associated with human temporal cortical architecture and Alzheimer’s pathology. bioRxiv. https://doi.org/10.1101/2021.07.07.451554
Chen WT, Lu A, Craessaerts K, Pavie B, Sala Frigerio C, Corthout N et al (2020) Spatial transcriptomics and in situ sequencing to study Alzheimer’s disease. Cell 182(976–991):e919. https://doi.org/10.1016/j.cell.2020.06.038
Cortes CJ, La Spada AR (2019) TFEB dysregulation as a driver of autophagy dysfunction in neurodegenerative disease: molecular mechanisms, cellular processes, and emerging therapeutic opportunities. Neurobiol Dis 122:83–93. https://doi.org/10.1016/j.nbd.2018.05.012
de Calignon A, Polydoro M, Suarez-Calvet M, William C, Adamowicz DH, Kopeikina KJ et al (2012) Propagation of tau pathology in a model of early Alzheimer’s disease. Neuron 73:685–697. https://doi.org/10.1016/j.neuron.2011.11.033
Delpech JC, Pathak D, Varghese M, Kalavai SV, Hays EC, Hof PR et al (2021) Wolframin-1-expressing neurons in the entorhinal cortex propagate tau to CA1 neurons and impair hippocampal memory in mice. Sci Transl Med 13:eabe8455. https://doi.org/10.1126/scitranslmed.abe8455
Fisher DW, Bennett DA, Dong H (2018) Sexual dimorphism in predisposition to Alzheimer’s disease. Neurobiol Aging 70:308–324. https://doi.org/10.1016/j.neurobiolaging.2018.04.004
Fonseca SG, Fukuma M, Lipson KL, Nguyen LX, Allen JR, Oka Y et al (2005) WFS1 is a novel component of the unfolded protein response and maintains homeostasis of the endoplasmic reticulum in pancreatic beta-cells. J Biol Chem 280:39609–39615. https://doi.org/10.1074/jbc.M507426200
Fonseca SG, Ishigaki S, Oslowski CM, Lu S, Lipson KL, Ghosh R et al (2010) Wolfram syndrome 1 gene negatively regulates ER stress signaling in rodent and human cells. J Clin Invest 120:744–755. https://doi.org/10.1172/JCI39678
Fredriksson S, Gullberg M, Jarvius J, Olsson C, Pietras K, Gustafsdottir SM et al (2002) Protein detection using proximity-dependent DNA ligation assays. Nat Biotechnol 20:473–477. https://doi.org/10.1038/nbt0502-473
Fu H, Hardy J, Duff KE (2018) Selective vulnerability in neurodegenerative diseases. Nat Neurosci 21:1350–1358. https://doi.org/10.1038/s41593-018-0221-2
Fu H, Hussaini SA, Wegmann S, Profaci C, Daniels JD, Herman M et al (2016) 3D visualization of the temporal and spatial spread of tau pathology reveals extensive sites of tau accumulation associated with neuronal loss and recognition memory deficit in aged tau transgenic mice. PLoS ONE 11:e0159463. https://doi.org/10.1371/journal.pone.0159463
Fu H, Possenti A, Freer R, Nakano Y, Hernandez Villegas NC, Tang M et al (2019) A tau homeostasis signature is linked with the cellular and regional vulnerability of excitatory neurons to tau pathology. Nat Neurosci 22:47–56. https://doi.org/10.1038/s41593-018-0298-7
Fu H, Rodriguez GA, Herman M, Emrani S, Nahmani E, Barrett G et al (2017) Tau pathology induces excitatory neuron loss, grid cell dysfunction, and spatial memory deficits reminiscent of early Alzheimer’s disease. Neuron 93(533–541):e535. https://doi.org/10.1016/j.neuron.2016.12.023
Fujita E, Kouroku Y, Isoai A, Kumagai H, Misutani A, Matsuda C et al (2007) Two endoplasmic reticulum-associated degradation (ERAD) systems for the novel variant of the mutant dysferlin: ubiquitin/proteasome ERAD(I) and autophagy/lysosome ERAD(II). Hum Mol Genet 16:618–629. https://doi.org/10.1093/hmg/ddm002
Grubman A, Chew G, Ouyang JF, Sun G, Choo XY, McLean C et al (2019) A single-cell atlas of entorhinal cortex from individuals with Alzheimer’s disease reveals cell-type-specific gene expression regulation. Nat Neurosci 22:2087–2097. https://doi.org/10.1038/s41593-019-0539-4
Guo JL, Narasimhan S, Changolkar L, He Z, Stieber A, Zhang B et al (2016) Unique pathological tau conformers from Alzheimer’s brains transmit tau pathology in nontransgenic mice. J Exp Med 213:2635–2654. https://doi.org/10.1084/jem.20160833
Halliday M, Radford H, Zents KAM, Molloy C, Moreno JA, Verity NC et al (2017) Repurposed drugs targeting eIF2α-P-mediated translational repression prevent neurodegeneration in mice. Brain 140:1768–1783. https://doi.org/10.1093/brain/awx074
Hetz C, Mollereau B (2014) Disturbance of endoplasmic reticulum proteostasis in neurodegenerative diseases. Nat Rev Neurosci 15:233–249. https://doi.org/10.1038/nrn3689
Iba M, McBride JD, Guo JL, Zhang B, Trojanowski JQ, Lee VM (2015) Tau pathology spread in PS19 tau transgenic mice following locus coeruleus (LC) injections of synthetic tau fibrils is determined by the LC’s afferent and efferent connections. Acta Neuropathol 130:349–362. https://doi.org/10.1007/s00401-015-1458-4
Ivask M, Hugill A, Koks S (2016) RNA-sequencing of WFS1-deficient pancreatic islets. Physiol Rep. https://doi.org/10.14814/phy2.12750
Ivask M, Volke V, Raasmaja A, Koks S (2021) High-fat diet associated sensitization to metabolic stress in Wfs1 heterozygous mice. Mol Genet Metab 134:203–211. https://doi.org/10.1016/j.ymgme.2021.07.002
Jack CR Jr, Bennett DA, Blennow K, Carrillo MC, Dunn B, Haeberlein SB et al (2018) NIA-AA research framework: toward a biological definition of Alzheimer’s disease. Alzheimers Dement 14:535–562. https://doi.org/10.1016/j.jalz.2018.02.018
Jack CR Jr, Holtzman DM (2013) Biomarker modeling of Alzheimer’s disease. Neuron 80:1347–1358. https://doi.org/10.1016/j.neuron.2013.12.003
Jack CR Jr, Knopman DS, Jagust WJ, Petersen RC, Weiner MW, Aisen PS et al (2013) Tracking pathophysiological processes in Alzheimer’s disease: an updated hypothetical model of dynamic biomarkers. Lancet Neurol 12:207–216. https://doi.org/10.1016/S1474-4422(12)70291-0
Jicha GA, Bowser R, Kazam IG, Davies P (1997) Alz-50 and MC-1, a new monoclonal antibody raised to paired helical filaments, recognize conformational epitopes on recombinant tau. J Neurosci Res 48:128–132. https://doi.org/10.1002/(sici)1097-4547(19970415)48:2%3c128::aid-jnr5%3e3.0.co;2-e
Jucker M, Walker LC (2018) Propagation and spread of pathogenic protein assemblies in neurodegenerative diseases. Nat Neurosci 21:1341–1349. https://doi.org/10.1038/s41593-018-0238-6
Kakiuchi C, Ishiwata M, Hayashi A, Kato T (2006) XBP1 induces WFS1 through an endoplasmic reticulum stress response element-like motif in SH-SY5Y cells. J Neurochem 97:545–555. https://doi.org/10.1111/j.1471-4159.2006.03772.x
Karran E, Mercken M, De Strooper B (2011) The amyloid cascade hypothesis for Alzheimer’s disease: an appraisal for the development of therapeutics. Nat Rev Drug Discov 10:698–712. https://doi.org/10.1038/nrd3505
Kawano J, Fu**aga R, Yamamoto-Hanada K, Oka Y, Tanizawa Y, Shinoda K (2009) Wolfram syndrome 1 (Wfs1) mRNA expression in the normal mouse brain during postnatal development. Neurosci Res 64:213–230. https://doi.org/10.1016/j.neures.2009.03.005
Kitamura T (2017) Driving and regulating temporal association learning coordinated by entorhinal-hippocampal network. Neurosci Res 121:1–6. https://doi.org/10.1016/j.neures.2017.04.005
Kitamura T, Pignatelli M, Suh J, Kohara K, Yoshiki A, Abe K et al (2014) Island cells control temporal association memory. Science 343:896–901. https://doi.org/10.1126/science.1244634
Koks S, Soomets U, Paya-Cano JL, Fernandes C, Luuk H, Plaas M et al (2009) Wfs1 gene deletion causes growth retardation in mice and interferes with the growth hormone pathway. Physiol Genomics 37:249–259. https://doi.org/10.1152/physiolgenomics.90407.2008
Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z et al (2016) Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44:W90-97. https://doi.org/10.1093/nar/gkw377
Lasagna-Reeves CA, de Haro M, Hao S, Park J, Rousseaux MW, Al-Ramahi I et al (2016) Reduction of Nuak1 decreases tau and reverses phenotypes in a tauopathy mouse model. Neuron 92:407–418. https://doi.org/10.1016/j.neuron.2016.09.022
Leng K, Li E, Eser R, Piergies A, Sit R, Tan M et al (2021) Molecular characterization of selectively vulnerable neurons in Alzheimer’s disease. Nat Neurosci 24:276–287. https://doi.org/10.1038/s41593-020-00764-7
Li L, Venkataraman L, Chen S, Fu H (2020) Function of WFS1 and WFS2 in the central nervous system: implications for Wolfram syndrome and Alzheimer’s disease. Neurosci Biobehav Rev 118:775–783. https://doi.org/10.1016/j.neubiorev.2020.09.011
Lie PPY, Yang DS, Stavrides P, Goulbourne CN, Zheng P, Mohan PS et al (2021) Post-Golgi carriers, not lysosomes, confer lysosomal properties to pre-degradative organelles in normal and dystrophic axons. Cell Rep 35:109034. https://doi.org/10.1016/j.celrep.2021.109034
Liu L, Drouet V, Wu JW, Witter MP, Small SA, Clelland C et al (2012) Trans-synaptic spread of tau pathology in vivo. PLoS ONE 7:e31302. https://doi.org/10.1371/journal.pone.0031302
Martini-Stoica H, Cole AL, Swartzlander DB, Chen F, Wan YW, Bajaj L et al (2018) TFEB enhances astroglial uptake of extracellular tau species and reduces tau spreading. J Exp Med 215:2355–2377. https://doi.org/10.1084/jem.20172158
Martini-Stoica H, Xu Y, Ballabio A, Zheng H (2016) The autophagy-lysosomal pathway in neurodegeneration: a TFEB perspective. Trends Neurosci 39:221–234. https://doi.org/10.1016/j.tins.2016.02.002
Mathys H, Davila-Velderrain J, Peng Z, Gao F, Mohammadi S, Young JZ et al (2019) Single-cell transcriptomic analysis of Alzheimer’s disease. Nature 570:332–337. https://doi.org/10.1038/s41586-019-1195-2
Meier S, Bell M, Lyons DN, Ingram A, Chen J, Gensel JC et al (2015) Identification of novel tau interactions with endoplasmic reticulum proteins in Alzheimer’s disease brain. J Alzheimers Dis 48:687–702. https://doi.org/10.3233/JAD-150298
Menzies FM, Fleming A, Rubinsztein DC (2015) Compromised autophagy and neurodegenerative diseases. Nat Rev Neurosci 16:345–357. https://doi.org/10.1038/nrn3961
Montine TJ, Phelps CH, Beach TG, Bigio EH, Cairns NJ, Dickson DW et al (2012) National Institute on Aging-Alzheimer’s Association guidelines for the neuropathologic assessment of Alzheimer’s disease: a practical approach. Acta Neuropathol 123:1–11. https://doi.org/10.1007/s00401-011-0910-3
Morrison JH, Hof PR (2007) Life and death of neurons in the aging cerebral cortex. Int Rev Neurobiol 81:41–57. https://doi.org/10.1016/S0074-7742(06)81004-4
Morrison JH, Hof PR (2002) Selective vulnerability of corticocortical and hippocampal circuits in aging and Alzheimer’s disease. Prog Brain Res 136:467–486. https://doi.org/10.1016/s0079-6123(02)36039-4
Moser EI, Kropff E, Moser MB (2008) Place cells, grid cells, and the brain’s spatial representation system. Annu Rev Neurosci 31:69–89. https://doi.org/10.1146/annurev.neuro.31.061307.090723
Nakashima A, Cheng SB, Kusabiraki T, Motomura K, Aoki A, Ushijima A et al (2019) Endoplasmic reticulum stress disrupts lysosomal homeostasis and induces blockade of autophagic flux in human trophoblasts. Sci Rep 9:11466. https://doi.org/10.1038/s41598-019-47607-5
Narasimhan S, Guo JL, Changolkar L, Stieber A, McBride JD, Silva LV et al (2017) Pathological tau strains from human brains recapitulate the diversity of tauopathies in nontransgenic mouse brain. J Neurosci 37:11406–11423. https://doi.org/10.1523/JNEUROSCI.1230-17.2017
Radford H, Moreno JA, Verity N, Halliday M, Mallucci GR (2015) PERK inhibition prevents tau-mediated neurodegeneration in a mouse model of frontotemporal dementia. Acta Neuropathol 130:633–642. https://doi.org/10.1007/s00401-015-1487-z
Rashid HO, Yadav RK, Kim HR, Chae HJ (2015) ER stress: autophagy induction, inhibition and selection. Autophagy 11:1956–1977. https://doi.org/10.1080/15548627.2015.1091141
Rauch JN, Luna G, Guzman E, Audouard M, Challis C, Sibih YE et al (2020) LRP1 is a master regulator of tau uptake and spread. Nature 580:381–385. https://doi.org/10.1038/s41586-020-2156-5
Riggs AC, Bernal-Mizrachi E, Ohsugi M, Wasson J, Fatrai S, Welling C et al (2005) Mice conditionally lacking the Wolfram gene in pancreatic islet beta cells exhibit diabetes as a result of enhanced endoplasmic reticulum stress and apoptosis. Diabetologia 48:2313–2321. https://doi.org/10.1007/s00125-005-1947-4
Sakakibara Y, Sekiya M, Fujisaki N, Quan X, Iijima KM (2018) Knockdown of wfs1, a fly homolog of Wolfram syndrome 1, in the nervous system increases susceptibility to age- and stress-induced neuronal dysfunction and degeneration in Drosophila. PLoS Genet 14:e1007196. https://doi.org/10.1371/journal.pgen.1007196
Sanders DW, Kaufman SK, DeVos SL, Sharma AM, Mirbaha H, Li A et al (2014) Distinct tau prion strains propagate in cells and mice and define different tauopathies. Neuron 82:1271–1288. https://doi.org/10.1016/j.neuron.2014.04.047
Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA et al (2009) A gene network regulating lysosomal biogenesis and function. Science 325:473–477. https://doi.org/10.1126/science.1174447
Scrivo A, Bourdenx M, Pampliega O, Cuervo AM (2018) Selective autophagy as a potential therapeutic target for neurodegenerative disorders. Lancet Neurol 17:802–815. https://doi.org/10.1016/S1474-4422(18)30238-2
Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S et al (2011) TFEB links autophagy to lysosomal biogenesis. Science 332:1429–1433. https://doi.org/10.1126/science.1204592
Stancu IC, Vasconcelos B, Terwel D, Dewachter I (2014) Models of beta-amyloid induced Tau-pathology: the long and “folded” road to understand the mechanism. Mol Neurodegener 9:51. https://doi.org/10.1186/1750-1326-9-51
Stranahan AM, Mattson MP (2010) Selective vulnerability of neurons in layer II of the entorhinal cortex during aging and Alzheimer’s disease. Neural Plast 2010:108190. https://doi.org/10.1155/2010/108190
Takeda K, Inoue H, Tanizawa Y, Matsuzaki Y, Oba J, Watanabe Y et al (2001) WFS1 (Wolfram syndrome 1) gene product: predominant subcellular localization to endoplasmic reticulum in cultured cells and neuronal expression in rat brain. Hum Mol Genet 10:477–484. https://doi.org/10.1093/hmg/10.5.477
Terasmaa A, Soomets U, Oflijan J, Punapart M, Hansen M, Matto V et al (2011) Wfs1 mutation makes mice sensitive to insulin-like effect of acute valproic acid and resistant to streptozocin. J Physiol Biochem 67:381–390. https://doi.org/10.1007/s13105-011-0088-0
Urano F (2016) Wolfram syndrome: diagnosis, management, and treatment. Curr Diab Rep 16:6. https://doi.org/10.1007/s11892-015-0702-6
Vonsattel JP, Del Amaya MP, Keller CE (2008) Twenty-first century brain banking. Processing brains for research: the Columbia University methods. Acta Neuropathol 115:509–532. https://doi.org/10.1007/s00401-007-0311-9
Xu Y, Du S, Marsh JA, Horie K, Sato C, Ballabio A et al (2020) TFEB regulates lysosomal exocytosis of tau and its loss of function exacerbates tau pathology and spreading. Mol Psychiatry. https://doi.org/10.1038/s41380-020-0738-0
Yamada T, Ishihara H, Tamura A, Takahashi R, Yamaguchi S, Takei D et al (2006) WFS1-deficiency increases endoplasmic reticulum stress, impairs cell cycle progression and triggers the apoptotic pathway specifically in pancreatic beta-cells. Hum Mol Genet 15:1600–1609. https://doi.org/10.1093/hmg/ddl081
Yamamoto A, Simonsen A (2011) The elimination of accumulated and aggregated proteins: a role for aggrephagy in neurodegeneration. Neurobiol Dis 43:17–28. https://doi.org/10.1016/j.nbd.2010.08.015
Yang DS, Lee JH, Nixon RA (2009) Monitoring autophagy in Alzheimer’s disease and related neurodegenerative diseases. Methods Enzymol 453:111–144. https://doi.org/10.1016/S0076-6879(08)04006-8
Yoshiyama Y, Higuchi M, Zhang B, Huang SM, Iwata N, Saido TC et al (2007) Synapse loss and microglial activation precede tangles in a P301S tauopathy mouse model. Neuron 53:337–351. https://doi.org/10.1016/j.neuron.2007.01.010
Yu G, Wang LG, Han Y, He QY (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16:284–287. https://doi.org/10.1089/omi.2011.0118
Acknowledgements
This work was supported by awards K01-AG056673, R56-AG066782-01 and R01-AG075092-01 (H.F.) from the National Institute on Aging of the National Institutes of Health and the award R01-GM131399 (Q.M.) from the National Institute of General Medical Sciences. The work was also supported by the award of AARF-17-505009 (H.F.) from the Alzheimer’s Association, the W81XWH1910309 (H.F.) from the Department of Defense, and the 10x Genomics 2021 Neuroscience Challenge award. Neurobehavioral tests were performed in the Department of Neuroscience Rodent Behavioral Core at Ohio State University, which was supported by NINDS P30NS04578. Some images were taken in the Department of Neuroscience Image Core, which was supported by P30 NS104177. The MSD assay was performed by the Clinical Research Center Analytical & Development Lab at The Ohio State University supported by Award Number UL1TR002733 from the National Center for Advancing Translational Sciences. We sincerely thank Marc Diamond for sharing the RD-P301S-YFP lentivector and DS9 tau cell line, Peter Davies for providing MC1, PHF1, and DA9 tau antibodies, and University of Tartu for sharing the Wfs1 knockout mice. We also thank Drs. Gail V.W. Johnson, Wai Haung Yu, Eric Klann and Andrea Tedeschi for helpful discussion. We thank the 10x Genomics technical support team for helpful discussions. We also thank Amanda Toland, Pearlly Yan, Tom Liu, Jennifer Mele from the Ohio State University and Amy Wetzel from the Nationwide Children’s Hospital for hel** with the RNA quality control and the sequencing. The RNA quality control was performed at NCI subsidized shared resource supported by The Ohio State University Comprehensive Cancer Center and the National Institutes of Health under grant number P30 CA016058. We thank Michael Rose and Rebecca Davis at The Ohio State University for preparing the human brain samples. Human de-identified brain tissues were kindly provided by the Banner Sun Health Research Institute Brain and Body Donation Program, supported by NIH grants U24-NS072026 and P30-AG19610 (TGB), the Arizona Department of Health Services (contract 211002, Arizona Alzheimer’s Research Center), the Arizona Biomedical Research Commission (contracts 4001, 0011, 05-901 and 1001 to the Arizona Parkinson's Disease Consortium) and the Michael J. Fox Foundation for Parkinson’s Research as well as the Buckeye Brain Bank and the Buckeye Biospecimen Repository at the Ohio State University, and the New York Brain Bank at Columbia University Irving Medical Center supported by the Taub Institute and NIH grants P50AG008702 and P30AG066462. This work used the high-performance computing infrastructure at the Ohio State University.
Author information
Authors and Affiliations
Contributions
HF designed and supervised the study, discussed the results, and wrote the paper. SC, DA, LL, and JL designed and performed experiments and analyzed the data and helped write the paper. YC generated all the pipelines for analyzing the 10 × Genomics Visium datasets. CW performed the single-nucleus RNA-Seq analysis. JF and ONK performed the behavioral tests. CM helped with the imaging data analysis. CNG performed the immuno-electron microscopy. YN, NCHV, and LV took part in the pilot studies. CB and MW helped with the PCR. GES, EB, XEF, LSH, JPV, DWS, and TGB prepared and characterized the human brain samples. T.W. performed the MSD assay. PP, TGB, QM, JK, SK, FU, and KED discussed and edited this paper.
Corresponding author
Ethics declarations
Conflict of interest
All authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chen, S., Acosta, D., Li, L. et al. Wolframin is a novel regulator of tau pathology and neurodegeneration. Acta Neuropathol 143, 547–569 (2022). https://doi.org/10.1007/s00401-022-02417-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00401-022-02417-4