Abstract
Key message
The major insight in this manuscript is that we identified a new flowering regulator, GmSOC1-like, which may participate in the initiation and maintenance of flowering in soybean.
Abstract
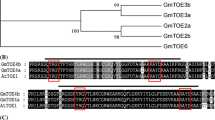
Flowering is pivotal for the reproductive behavior of plants, and it is regulated by complex and coordinated genetic networks that are fine-tuned by endogenous cues and environmental signals. To better understand the molecular basis of flowering regulation in soybean, we isolated GmSOC1 and GmSOC1-like, two putative soybean orthologs for the Arabidopsis SUPPRESSOR OF OVEREXPRESSION OF CO1/AGAMOUS-LIKE 20 (SOC1/AGL20). The expression pattern of GmSOC1-like was analyzed by qRT-PCR in Zigongdongdou, a photoperiod-sensitive soybean cultivar. GmSOC1-like was widely expressed at different levels in most organs of the soybean, with the highest expression in the shoot apex during the early stage of floral transition. In addition, its expression showed a circadian rhythm pattern, with the highest expression at midnight under short-day (SD) condition. Intriguingly, GmSOC1-like was induced 4 days earlier than GmSOC1 during flowering transition in SD, suggesting that GmSOC1 and GmSOC1-like expression might be differentially regulated. However, under long-day (LD) condition, the expression of GmSOC1 and GmSOC1-like decreased gradually in the shoot apex of Zigongdongdou, which is in accordance with the fact that Zigongdongdou maintains vegetative growth in LD. In addition, overexpression of GmSOC1-like stimulated the flowering of Lotus corniculatus cv. supperroot plants. In conclusion, the results of this study indicate that GmSOC1-like may act as a flowering inducer in soybean.







Similar content being viewed by others
References
Abe J, Xu D, Miyano A et al (2003) Photoperiod-insensitive Japanese soybean landraces differ at two maturity loci. Crop Sci 43:1300–1304
Akashi R, Kawano T, Hashiguchi M et al (2003) Super roots in Lotus corniculatus: a unique tissue culture and regeneration system in a legume species. Plant Soil 255:27–33
Alvarez-Buylla ER, Liljegren SJ, Pelaz S et al (2000) MADS-box gene evolution beyond flowers: expression in pollen, endosperm, guard cells, roots and trichomes. Plant J 24:457–466
Amasino RM (2005) Vernalization and flowering time. Curr Opin Biotechnol 16:154–158
Balasubramanian S, Sureshkumar S, Lempe J et al (2006) Potent induction of Arabidopsis thaliana flowering by elevated growth temperature. PLoS Genet 2:e106
Bashirullah A, Cooperstock RL, Lipshitz HD (2001) Spatial and temporal control of RNA stability. Proc Natl Acad Sci USA 98:7025–7028
Bernard RL (1971) Two major genes for time of flowering and maturity in soybeans. Crop Sci 11:242–244
Blazquez MA, Ahn JH, Weigel D (2003) A thermosensory pathway controlling flowering time in Arabidopsis thaliana. Nat Genet 33:168–171
Bonato ER, Vello NA (1999) E6, a dominant gene conditioning early flowering and maturity in soybeans. Genet Mol Biol 22:229–232
Buzzell RI (1971) Inheritance of a soybean flowering response to fluorescent-daylength conditions. Can J Genet Cytol 13:703–707
Buzzell RI, Voldeng HD (1980) Inheritance of insensitivity to long day length. Soybean Genet Newsl 7:26–29
Cerdán PD, Chory J (2003) Regulation of flowering time by light quality. Nature 423:881–885
Chi Y, Huang F, Liu H et al (2011) An APETALA1-like gene of soybean regulates flowering time and specifies floral organs. J Plant Physiol 168:2251–2259
Choi K, Kim J, Hwang HJ et al (2011) The FRIGIDA complex activates transcription of FLC, a strong flowering repressor in Arabidopsis, by recruiting chromatin modification factors. Plant Cell 23:289–303
Cober ER, Morrison MJ (2010) Regulation of seed yield and agronomic characters by photoperiod sensitivity and growth habit genes in soybean. Theor Appl Genet 120:1005–1012
Cober ER, Tanner JW, Voldeng HD (1996a) Genetic control of photoperiod response in early maturing near-isogenic soybean lines. Crop Sci 36:601–605
Cober ER, Tanner JW, Voldeng HD (1996b) Soybean photoperiod-sensitivity loci respond differentially to light quality. Crop Sci 36:606–610
Cober ER, Molnar SJ, Charette M et al (2010) A new locus for early maturity in soybean. Crop Sci 50:524–527
Cseke LJ, Zheng J, Podila GK (2003) Characterization of PTM5 in aspen trees: a MADS-box gene expressed during woody vascular development. Gene 318:55–67
Decroocq V, Zhu X, Kauffman M et al (1999) A TM3-like MADS-box gene from Eucalyptus expressed in both vegetative and reproductive tissues. Gene 228:155–160
Deng W, Ying H, Helliwell CA et al (2011) FLOWERING LOCUS C (FLC) regulates development pathways throughout the life cycle of Arabidopsis. Proc Natl Acad Sci USA 108:6680–6685
Ferrario S, Busscher J, Franken J et al (2004) Ectopic expression of the petunia MADS box gene UNSHAVEN accelerates flowering and confers leaflike characteristics to floral organs in a dominant-negative manner. Plant Cell 16:1490–1505
Gregis V, Sessa A, Dorca-Fornell C et al (2009) The Arabidopsis floral meristem identity genes AP1, AGL24 and SVP directly repress class B and C floral homeotic genes. Plant J 60:626–637
Han T, Wang J (1995) Studies on the post-flowering photoperiodic responses in soybean. Acta Bot Sin 37:863–869
Han T, Gai J, Wang J et al (1998) Discovery of flowering reversions in soybean plant. Acta Agron Sinica 24:168–171
Hayama R, Coupland G (2003) Shedding light on the circadian clock and the photoperiodic control of flowering. Curr Opin Plant Biol 6:13–19
Hepworth SR, Valverde F, Ravenscroft D et al (2002) Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. EMBO J 21:4327–4337
Immink RGH, Angenent GC (2002) Transcription factors do it together: the hows and whys of studying protein–protein interactions. Trends Plant Sci 7:531–534
Jang S, Marchal V, Panigrahi KCS et al (2008) Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J 27:1277–1288
Jansen RP (2001) mRNA localization: message on the move. Nat Rev Mol Cell Biol 2:247–256
Jian B, Hou W, Wu C et al (2009) Agrobacterium rhizogenes-mediated transformation of Superroot-derived Lotus corniculatus plants: a valuable tool for functional genomics. BMC Plant Biol 9:78
Kelley LA, Sternberg MJE (2009) Protein structure prediction on the web: a case study using the Phyre server. Nat Protoc 4:363–371
Kong F, Liu B, **a Z et al (2010) Two coordinately regulated homologs of FLOWERING LOCUS T are involved in the control of photoperiodic flowering in soybean. Plant Physiol 154:1220–1231
Lee J, Lee I (2010) Regulation and function of SOC1, a flowering pathway integrator. J Exp Bot 61:2247–2254
Lee H, Suh SS, Park E et al (2000) The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev 14:2366–2376
Lee S, Kim J, Han JJ et al (2004) Functional analyses of the flowering time gene OsMADS50, the putative SUPPRESSOR OF OVEREXPRESSION OF CO 1/AGAMOUS-LIKE 20 (SOC1/AGL20) ortholog in rice. Plant J 38:754–764
Lee J, Oh M, Park H et al (2008) SOC1 translocated to the nucleus by interaction with AGL24 directly regulates LEAFY. Plant J 55:832–843
Li X, Wu C, Ma Q et al (2005) Morphology and anatomy of the differentiation of flower bud and the process of flowering reversion in soybean cv. Zigongdongdou. Acta Agron Sin 31:1437–1442
Li HJ, Xue Y, Jia DJ et al (2011) POD1 regulates pollen tube guidance in response to micropylar female signaling and acts in early embryo patterning in Arabidopsis. Plant Cell 23:3288–3302
Liu C, Chen H, Er HL et al (2008) Direct interaction of AGL24 and SOC1 integrates flowering signals in Arabidopsis. Development 135:1481–1491
McBlain BA, Bernard RL, Cremeens CR et al (1987) A procedure to identify genes affecting maturity using soybean isoline testers. Crop Sci 27:1127–1132
Michaels SD, Ditta G, Gustafson-Brown C et al (2003) AGL24 acts as a promoter of flowering in Arabidopsis and is positively regulated by vernalization. Plant J 33:867–874
Mignone F, Gissi C, Liuni S et al (2002) Untranslated regions of mRNAs. Genome Biol 3:0004.1–0004.10
Moon J, Suh SS, Lee H et al (2003) The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J 35:613–623
Mouradov A, Cremer F, Coupland G (2002) Control of flowering time interacting pathways as a basis for diversity. Plant Cell 14(suppl 1):S111–S130
Ng M, Yanofsky MF (2001) Function and evolution of the plant MADS-box gene family. Nat Rev Genet 2:186–195
Pelaz S, Ditta GS, Baumann E et al (2000) B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405:200–203
Pellegrini L, Tan S, Richmond TJ (1995) Structure of serum response factor core bound to DNA. Nature 376:490–498
Riechmann JL, Meyerowitz EM (1997) MADS domain proteins in plant development. Biol Chem 378:1079–1101
Riechmann JL, Ratcliffe OJ (2000) A genomic perspective on plant transcription factors. Curr Opin Plant Biol 3:423–434
Riechmann JL, Wang M, Meyerowitz EM (1996) DNA-binding properties of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA and AGAMOUS. Nucleic Acids Res 24:3134–3141
Samach A, Onouchi H, Gold SE et al (2000) Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288:1613–1616
Santelli E, Richmond TJ (2000) Crystal structure of MEF2A core bound to DNA at 1.5 Å resolution. J Mol Biol 297:437–449
Searle I, He Y, Turck F et al (2006) The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev 20:898–912
Seo E, Lee H, Jeon J et al (2009) Crosstalk between cold response and flowering in Arabidopsis is mediated through the flowering-time gene SOC1 and its upstream negative regulator FLC. Plant Cell 21:3185–3197
Shore P, Sharrocks AD (1995) The MADS-box family of transcription factors. Eur J Biochem 229:1–13
Simpson GG, Dean C (2002) Arabidopsis, the Rosetta stone of flowering time? Science 296:285–289
Sridhar VV, Surendrarao A, Liu Z et al (2006) APETALA1 and SEPALLATA3 interact with SEUSS to mediate transcription repression during flower development. Development 133:3159–3166
Suárez-López P, Wheatley K, Robson F et al (2001) CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410:1116–1120
Sun H, Jia Z, Cao D et al (2011) GmFT2a, a soybean homolog of FLOWERING LOCUS T, is involved in flowering transition and maintenance. PLoS ONE 6:e29238
Sung ZR, Chen L, Moon YH et al (2003) Mechanisms of floral repression in Arabidopsis. Curr Opin Plant Biol 6:29–35
Tadege M, Sheldon CC, Helliwell CA et al (2003) Reciprocal control of flowering time by OsSOC1 in transgenic Arabidopsis and by FLC in transgenic rice. Plant Biotech J 1:361–369
Tan S, Richmond TJ (1998) Crystal structure of the yeast MATalpha2/MCM1/DNA ternary complex. Nature 391:660–666
Tan FC, Swain SM (2007) Functional characterization of AP3, SOC1 and WUS homologues from citrus (Citrus sinensis). Physiol Plant 131:481–495
Theissen G, Becker A, Di Rosa A et al (2000) A short history of MADS-box genes in plants. Plant Mol Biol 42:115–149
Tiwari SB, Shen Y, Chang HC et al (2010) The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytol 187:57–66
van Der Velden AW, Thomas AAM (1999) The role of the 5′ untranslated region of an mRNA in translation regulation during development. Int J Biochem Cell Biol 31:87–106
Verelst W, Saedler H, Münster T (2007) MIKC* MADS-protein complexes bind motifs enriched in the proximal region of late pollen-specific Arabidopsis promoters. Plant Physiol 143:447–460
Wang JW, Czech B, Weigel D (2009) miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138:738–749
Watanabe S, **a Z, Hideshima R et al (2011) A map-based cloning strategy employing a residual heterozygous line reveals that the GIGANTEA gene is involved in soybean maturity and flowering. Genetics 188:395–407
Wigge PA, Kim MC, Jaeger KE et al (2005) Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309:1056–1059
Wilkie GS, Dickson KS, Gray NK (2003) Regulation of mRNA translation by 5′- and 3′-UTR-binding factors. Trends Biochem Sci 28:182–188
Wu C, Ma Q, Yam K-M et al (2006) In situ expression of the GmNMH7 gene is photoperiod-dependent in a unique soybean (Glycine max [L.] Merr.) flowering reversion system. Planta 223:725–735
Wu G, Park MY, Conway SR et al (2009) The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138:750–759
**a Z, Watanabe S, Yamada T et al (2012) Positional cloning and characterization reveal the molecular basis for soybean maturity locus E1 that regulates photoperiodic flowering. Proc Natl Acad Sci USA 109:E2155–E2164
**e ZM, Zou HF, Lei G et al (2009) Soybean trihelix transcription factors GmGT-2A and GmGT-2B improve plant tolerance to abiotic stresses in transgenic Arabidopsis. PLoS ONE 4:e6898
Yoo SK, Chung KS, Kim J et al (2005) CONSTANS activates SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 through FLOWERING LOCUS T to promote flowering in Arabidopsis. Plant Physiol 139:770–778
Zhang Q, Li H, Li R et al (2008) Association of the circadian rhythmic expression of GmCRY1a with a latitudinal cline in photoperiodic flowering of soybean. Proc Natl Acad Sci USA 105:21028–21033
Zhong X, Dai X, Xv J et al (2012) Cloning and expression analysis of GmGAL1, SOC1 homolog gene in soybean. Mol Bio Rep 39:6967–6974
Acknowledgments
We thank Dr. Ryo Akashi at the Biological Resource Center in Lotus and Glycine (University of Miyazaki, Japan) for providing the Superroot culture of L. corniculatus; Professor Peter Gresshoff (University of Queensland, Australia) for A. rhizogenes strain K599 and the binary vector pGFPGUSPlus; and Dr. Hongbo Sun (Minzu University of China) for stimulating discussion on experimental design. This work was supported by the National Program on Key Basic Research Project (973 Program) of China (2009CB118400) and Natural Science Foundation of China (30471054).
Conflict of interest
The authors declare that they have no conflict interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by K. Chong.
X. Na and B. Jian contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Na, X., Jian, B., Yao, W. et al. Cloning and functional analysis of the flowering gene GmSOC1-like, a putative SUPPRESSOR OF OVEREXPRESSION CO1/AGAMOUS-LIKE 20 (SOC1/AGL20) ortholog in soybean. Plant Cell Rep 32, 1219–1229 (2013). https://doi.org/10.1007/s00299-013-1419-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-013-1419-0