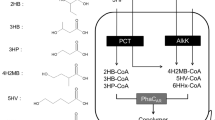
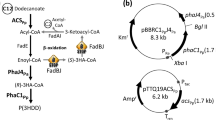
Abstract
Bacterially produced biodegradable polyhydroxyalkanoates (PHAs) with versatile properties can be achieved using different PHA synthases (PhaCs). This work aims to expand the diversity of known PhaCs via functional metagenomics and demonstrates the use of these novel enzymes in PHA production. Complementation of a PHA synthesis-deficient Pseudomonas putida strain with a soil metagenomic cosmid library retrieved 27 clones expressing either class I, class II, or unclassified PHA synthases, and many did not have close sequence matches to known PhaCs. The composition of PHA produced by these clones was dependent on both the supplied growth substrates and the nature of the PHA synthase, with various combinations of short-chain-length (SCL) and medium-chain-length (MCL) PHA. These data demonstrate the ability to isolate diverse genes for PHA synthesis by functional metagenomics and their use for the production of a variety of PHA polymer and copolymer mixtures.




Similar content being viewed by others
References
Bagdasarian M, Lurz R, Rückert B, Franklin FC, Bagdasarian MM, Frey J, Timmis KN (1981) Specific purpose plasmid cloning vectors II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene 16:237–247
Cheema S, Bassas-Galia M, Sarma PM, Lal B, Arias S (2012) Exploiting metagenomic diversity for novel polyhydroxyalkanoate synthases: production of a terpolymer poly(3-hydroxybutyrate-co-3-hydroxyhexanoate-co-3-hydroxyoctanoate) with a recombinant Pseudomonas putida strain. Bioresour Technol 103:322–328. doi:10.1016/j.biortech.2011.09.098
Chen Y-J, Tsai P-C, Hsu C-H, Lee C-Y (2014) Critical residues of class II PHA synthase for expanding the substrate specificity and enhancing the biosynthesis of polyhydroxyalkanoate. Enzym Microb Technol 56:60–66. doi:10.1016/j.enzmictec.2014.01.005
Cheng J, Pinnell L, Engel K, Neufeld JD, Charles TC (2014) Versatile broad-host-range cosmids for construction of high quality metagenomic libraries. J Microbiol Methods 99:27–34. doi:10.1016/j.mimet.2014.01.015
de Eugenio LI, Escapa IF, Morales V, Dinjaski N, Galàn B, García JL, Prieto MA (2010) The turnover of medium-chain-length polyhydroxyalkanoates in Pseudomonas putida KT2442 and the fundamental role of PhaZ depolymerase for the metabolic balance. Environ Microbiol 12:207–221. doi:10.1111/j.1462-2920.2009.02061.x
Edgar RC (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinform 5:113–119. doi:10.1186/1471-2105-5-113
Escapa IF, Morales V, Martino VP, Pollet E, Avérous L, García JL, Prieto MA (2011) Disruption of β-oxidation pathway in Pseudomonas putida KT2442 to produce new functionalized PHAs with thioester groups. Appl Microbiol Biotechnol 89:1583–1598. doi:10.1007/s00253-011-3099-4
Fellay R, Frey J, Krisch H (1987) Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of gram-negative bacteria. Gene 52:147–154
Finan TM, Kunkel B, DeVos GF, Signer ER (1986) Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J Bacteriol 167:66–72
Foong CP, Lau N-S, Deguchi S, Toyofuku T, Taylor TD, Sudesh K, Matsui M (2014) Whole genome amplification approach reveals novel polyhydroxyalkanoate synthases (PhaCs) from Japan trench and nankai trough seawater. BMC Microbiol 14:e11836. doi:10.1186/s12866-014-0318-z
Fukui T, Doi Y (1997) Cloning and analysis of the poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) biosynthesis genes of Aeromonas caviae. J Bacteriol 179:4821–4830. doi:10.1186/s12866-014-0318-z
Fukui T, Shiomi N, Doi Y (1998) Expression and characterization of (R)-specific enoyl coenzyme A hydratase involved in polyhydroxyalkanoate biosynthesis by Aeromonas caviae. J Bacteriol 180:667–673
Hoppensack A, Rehm BH, Steinbüchel A (1999) Analysis of 4-phosphopantetheinylation of polyhydroxybutyrate synthase from Ralstonia eutropha: generation of beta-alanine auxotrophic Tn5 mutants and cloning of the panD gene region. J Bacteriol 181:1429–1435
Jia Y, Kappock TJ, Frick T, Sinskey AJ, Stubbe J (2000) Lipases provide a new mechanistic model for polyhydroxybutyrate (PHB) synthases: characterization of the functional residues in Chromatium vinosum PHB synthase. Biochemistry 39:3927–3936. doi:10.1021/bi9928086
Jones JDG, Gutterson N (1987) An efficient mobilizable cosmid vector, pRK7813, and its use in a rapid method for marker exchange in Pseudomonas fluorescens strain HV37a. Gene 61:299–306
Kawashima Y, Cheng W, Mifune J, Orita I, Nakamura S, Fukui T (2012) Characterization and functional analyses of R-specific enoyl coenzyme a hydratases in polyhydroxyalkanoate-producing Ralstonia eutropha. Appl Environ Microbiol 78:493–502. doi:10.1128/AEM.06937-11
Keshavarz T, Roy I (2010) Polyhydroxyalkanoates: bioplastics with a green agenda. Curr Opin Microbiol 13:321–326. doi:10.1016/j.mib.2010.02.006
Lam KN, Hall MW, Engel K, Vey G, Cheng J, Neufeld JD, Charles TC (2014) Evaluation of a pooled strategy for high-throughput sequencing of cosmid clones from metagenomic libraries. PLoS One 9:e98968. doi:10.1371/journal.pone.0098968
Matsumoto K, Takase K, Aoki E, Doi Y, Taguchi S (2005) Synergistic effects of Glu130Asp substitution in the type II polyhydroxyalkanoate (PHA) synthase: enhancement of PHA production and alteration of polymer molecular weight. Biomacromolecules 6:99–104. doi:10.1021/bm049650b
Matsusaki H, Manji S, Taguchi K, Kato M, Fukui T, Doi Y (1998) Cloning and molecular analysis of the poly(3-hydroxybutyrate) and poly(3-hydroxybutyrate-co-3-hydroxyalkanoate) biosynthesis genes in Pseudomonas sp. strain 61-3. J Bacteriol 180:6459–6467
Meade HM, Long SR, Ruvkun GB, Brown SE, Ausubel FM (1982) Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon mutagenesis. J Bacteriol 149:114–122
Meng D-C, Shen R, Yao H, Chen J-C, Wu Q, Chen G-Q (2014) Engineering the diversity of polyesters. Curr Opin Biotechnol 29:24–33. doi:10.1016/j.copbio.2014.02.013
Noda I, Green PR, Satkowski MM, Schechtman LA (2005) Preparation and properties of a novel class of polyhydroxyalkanoate copolymers. Biomacromolecules 6:580–586. doi:10.1021/bm049472m
Park SJ, Kim TW, Kim MK, Lee SY, Lim S-C (2012) Advanced bacterial polyhydroxyalkanoates: towards a versatile and sustainable platform for unnatural tailor-made polyesters. Biotechnol Adv 30:1196–1206. doi:10.1016/j.biotechadv.2011.11.007
Pärnänen K, Karkman A, Virta M, Eronen-Rasimus E, Kaartokallio H (2015) Discovery of bacterial polyhydroxyalkanoate synthase (PhaC)-encoding genes from seasonal Baltic Sea ice and cold estuarine waters. Extremophiles 19:197–206. doi:10.1007/s00792-014-0699-9
Patil KR, Roune L, McHardy AC (2012) The PhyloPythiaS web server for taxonomic assignment of metagenome sequences. PLoS One 7:e38581. doi:10.1371/journal.pone.0038581
Quandt J, Hynes MF (1993) Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127:15–21. doi:10.1016/0378-1119(93)90611-6
Rehm BHA (2003) Polyester synthases: natural catalysts for plastics. Biochem J 376:15–33. doi:10.1042/BJ20031254
Sato S, Kanazawa H, Tsuge T (2011) Expression and characterization of (R)-specific enoyl coenzyme a hydratases making a channeling route to polyhydroxyalkanoate biosynthesis in Pseudomonas putida. Appl Microbiol Biotechnol 90:951–959. doi:10.1007/s00253-011-3150-5
Schallmey M, Ly A, Wang C, Meglei G, Voget S, Streit WR, Driscoll BT, Charles TC (2011) Harvesting of novel polyhydroxyalkanaote (PHA) synthase encoding genes from a soil metagenome library using phenotypic screening. FEMS Microbiol Lett 321:150–156. doi:10.1111/j.1574-6968.2011.02324.x
Sheu DS, Wang YT, Lee CY (2000) Rapid detection of polyhydroxyalkanoate-accumulating bacteria isolated from the environment by colony PCR. Microbiology 146:2019–2025. doi:10.1099/00221287-146-8-2019
Shozui F, Sun J, Song Y, Yamada M, Sakai K, Matsumoto K, Takase K, Taguchi S (2010) A new beneficial mutation in Pseudomonas sp. 61-3 polyhydroxyalkanoate (PHA) synthase for enhanced cellular content of 3-hydroxybutyrate-based PHA explored using its enzyme homolog as a mutation template. Biosci Biotechnol Biochem 74:1710–1712. doi:10.1271/bbb.100224
Simon C, Daniel R (2011) Metagenomic analyses: past and future trends. Appl Environ Microbiol 77:1153–1161. doi:10.1128/AEM.02345-10
Steinbüchel A, Lütke-Eversloh T (2003) Metabolic engineering and pathway construction for biotechnological production of relevant polyhydroxyalkanoates in microorganisms. Biochem Eng J 16:81–96. doi:10.1016/S1369-703X(03)00036-6
Tai YT, Foong CP, Najimudin N, Sudesh K (2015) Discovery of a new polyhydroxyalkanoate synthase from limestone soil through metagenomic approach. J Biosci Bioeng. doi:10.1016/j.jbiosc.2015.08.008
Takase K, Taguchi S, Doi Y (2003) Enhanced synthesis of poly(3-hydroxybutyrate) in recombinant Escherichia coli by means of error-prone PCR mutagenesis, saturation mutagenesis, and in vitro recombination of the type II polyhydroxyalkanoate synthase gene. J Biochem 133:139–145
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi:10.1093/molbev/mst197
Tortajada M, da Silva LF, Prieto MA (2013) Second-generation functionalized medium-chain-length polyhydroxyalkanoates: the gateway to high-value bioplastic applications. Int Microbiol 16:1–15. doi:10.2436/20.1501.01.175
Tripathi L, Wu L-P, Dechuan M, Chen J, Wu Q, Chen G-Q (2013) Pseudomonas putida KT2442 as a platform for the biosynthesis of polyhydroxyalkanoates with adjustable monomer contents and compositions. Bioresour Technol 142:225–231. doi:10.1016/j.biortech.2013.05.027
Verlinden RAJ, Hill DJ, Kenward MA, Williams CD, Radecka I (2007) Bacterial synthesis of biodegradable polyhydroxyalkanoates. J Appl Microbiol 102:1437–1449. doi:10.1111/j.1365-2672.2007.03335.x
Wu H-A, Sheu D-S, Lee C-Y (2003) Rapid differentiation between short-chain-length and medium-chain-length polyhydroxyalkanoate-accumulating bacteria with spectrofluorometry. J Microbiol Methods 53:131–135. doi:10.1016/S0167-7012(02)00232-4
Zhu W, Lomsadze A, Borodovsky M (2010) Ab initio gene identification in metagenomic sequences. Nucleic Acids Res 38:e132–e132. doi:10.1093/nar/gkq275
Acknowledgments
We are very grateful to Ricardo Nordeste for assisting with GC-MS analysis of PHA. We wish to thank Dr. Bruce Ramsay (Polyferm Canada) for providing PHA standards and Dr. Josh Neufeld for use of the FilterMax F5 Multi-mode microplate reader.
Authors’s contributions
JC and TCC conceived and designed this study, carried out all the experiments, and prepared the manuscript. Both authors read and approved the final manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This work was financially supported by a New Directions grant from Ontario Ministry of Agriculture, Food, and Rural Affairs (award number 381646-09), by a Strategic Projects grant from the Natural Sciences and Engineering Research Council of Canada (award number ND2012-1679), and by Genome Canada, through the Applied Genomics Research in Bioproducts or Crops (ABC) program for the grant titled “Microbial Genomics for Biofuels and Co-Products from Biorefining Processes” awarded to David Levin and Richard Sparling.
Conflict of interest
Trevor Charles received partial financial support for presenting some of this material at the Society for Industrial Microbiology and Biotechnology Annual Meeting in 2014 and 2015. Jiujun Cheng declares no conflict of interest.
Ethical statement
The authors certify that this manuscript has not been published previously, and not under consideration for publication elsewhere, in whole or in part. No data have been fabricated or manipulated (including images), and no data, text, or theories by others are presented as if they were the authors’ own. Consent to submit has been received explicitly from all the authors listed. And authors whose names appear on the submission have contributed sufficiently to the scientific work and therefore share collective responsibility and accountability for the results. This article does not contain any studies with human participants or animals performed by any of the authors.
Electronic supplementary material
ESM 1
(PDF 621 kb)
Rights and permissions
About this article
Cite this article
Cheng, J., Charles, T.C. Novel polyhydroxyalkanoate copolymers produced in Pseudomonas putida by metagenomic polyhydroxyalkanoate synthases. Appl Microbiol Biotechnol 100, 7611–7627 (2016). https://doi.org/10.1007/s00253-016-7666-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-016-7666-6