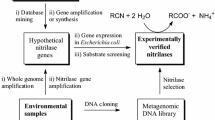
Abstract
The aim of this study was to discover new nitrilases with useful activities, especially towards dinitriles that are precursors of high-value cyano acids. Genes coding for putative nitrilases of different origins (fungal, plant, or bacterial) with moderate similarities to known nitrilases were selected by mining the GenBank database, synthesized artificially and expressed in Escherichia coli. The enzymes were purified, examined for their substrate specificities, and classified into subtypes (aromatic nitrilase, arylacetonitrilase, aliphatic nitrilase, cyanide hydratase) which were largely in accordance with those predicted from bioinformatic analysis. The catalytic potential of the nitrilases for dinitriles was examined with cyanophenyl acetonitriles, phenylenediacetonitriles, and fumaronitrile. The nitrilase activities and selectivities for dinitriles and the reaction products (cyano acid, cyano amide, diacid) depended on the enzyme subtype. At a preparative scale, all the examined dinitriles were hydrolyzed into cyano acids and fumaronitrile was converted to cyano amide using E. coli cells producing arylacetonitrilases and an aromatic nitrilase, respectively.

Similar content being viewed by others
References
Asano Y (2002) Overview of screening for new microbial catalysts and their uses in organic synthesis—selection and optimization of biocatalysts. J Biotechnol 94:65–72. doi:10.1016/S0168-1656(01)00419-9
Banerjee A, Dubey S, Kaul P, Barse B, Piotrowski M, Banerjee UC (2009) Enantioselective nitrilase from Pseudomonas putida: cloning, heterologous expression, and bioreactor studies. Mol Biotechnol 41:35–41. doi:10.1007/s12033-008-9094-z
Basile LJ, Willson RC, Sewell BT, Benedik MJ (2008) Genome mining of cyanide degrading nitrilases from filamentous fungi. Appl Microbiol Biotechnol 80:427–435. doi:10.1007/s00253-008-1559-2
Bayer S, Birkemeyer C, Ballschmiter M (2011) A nitrilase from a metagenomic library acts regioselectively on aliphatic dinitriles. Appl Environ Microbiol 89:91–98. doi:10.1007/s00253-010-2831-9
DeSantis G, Wong K, Farwell B, Chatman K, Zhu Z, Tomlinson G, Huang H, Tan X, Bibbs L, Chen P, Kretz K, Burk MJ (2003) Creation of a productive, highly enantioselective nitrilase through gene site saturation mutagenesis (GSSM). J Am Chem Soc 125:11476–11477. doi:10.1021/ja035742h
Duan YT, Yao PY, Ren J, Han C, Li Q, Yuan J, Feng JH, Wu QQ, Zhu DM (2014) Biocatalytic desymmetrization of 3-substituted glutaronitriles by nitrilases. A convenient chemoenzymatic access to optically active (S)-pregabalin and (R)-baclofen. Sci China Chem 57:1164–1171. doi:10.1007/s11426-014-5139-2
Effenberger F, Osswald S (2001) Selective hydrolysis of aliphatic dinitriles to monocarboxylic acids by a nitrilase from Arabidopsis thaliana. Synthesis-Stuttgart:1866–1872
Gökce H, Bahceli S (2011) Quantum chemical computations of 1,3-phenylenediacetic acid. Spectrochim Acta, Part A 78:803–808. doi:10.1016/j.saa.2010.12.031
Heinemann U, Engels D, Bürger S, Kiziak C, Mattes R, Stolz A (2003) Cloning of a nitrilase gene from the cyanobacterium Synechocystis sp. strain PCC6803 and heterologous expression and characterization of the encoded protein. Appl Environ Microbiol 69:4359–4366. doi:10.1128/AEM.69.8.4359-4366.2003
Hoyle AJ, Bunch AW, Knowles CJ (1998) The nitrilases of Rhodococcus rhodochrous NCIMB 11216. Enzym Microb Technol 23:475–482. doi:10.1016/S0141-0229(98)00076-3
Chauhan S, Wu S, Blumerman S, Fallon RD, Gavagan JE, DiCosimo R, Payne MS (2003) Purification, cloning, sequencing and over-expression in Escherichia coli of a regioselective aliphatic nitrilase from Acidovorax facilis 72W. Appl Microbiol Biotechnol 61:118–122. doi:10.1007/s00253-002-1192-4
Kaplan O, Vejvoda V, Plíhal O, Pompach P, Kavan D, Bojarová P, Bezouška K, Macková M, Cantarella M, Jirků V, Křen V, Martínková L (2006) Purification and characterization of a nitrilase from Aspergillus niger K10. Appl Microbiol Biotechnol 73:567–575. doi:10.1007/s00253-006-0503-6
Kaplan O, Veselá AB, Petříčková A, Pasquarelli F, Pičmanová M, Rinágelová A, Bhalla TC, Pátek M, Martínková L (2013a) A comparative study of nitrilases identified by genome mining. Mol Biotechnol 54:996–1003. doi:10.1007/s12033-013-9656-6
Kaplan O, Vejvoda V, Plíhal O, Pompach P, Kavan D, Bojarová P, Bezouška K, Macková M, Cantarella M, Jirků V, Křen V, Martínková L (2013b) Erratum to: purification and characterization of a nitrilase from Aspergillus niger K10 (vol 73, pg 567(2006). Appl Microbiol Biotechnol 97:3745–3746. doi:10.1007/s00253-013-4743-y
Kiziak C, Conradt D, Stolz A, Mattes R, Klein J (2005) Nitrilase from Pseudomonas fluorescens EBC191: cloning and heterologous expression of the gene and biochemical characterization of the recombinant enzyme. Microbiology 151:3639–3648. doi:10.1099/mic.0.28246-0
Kriechbaumer V, Park WJ, Piotrowski M, Meeley RB, Gierl A, Glawischnig E (2007) Maize nitrilases have a dual role in auxin homeostasis and β-cyanoalanine hydrolysis. J Exp Bot 58:4225–4233. doi:10.1093/jxb/erm279
Liu Z-Q, Zhang X-H, Xue Y-P, Xu M, Zheng Y-G (2014) Improvement of Alcaligenes faecalis nitrilase by gene site saturation mutagenesis and its application in stereospecific biosynthesis of (R)‑(−)-mandelic acid. Agric Food Chem 6:4685–4694. doi:10.1021/jf405683f
Martínková L, Křen V (2010) Biotransformations with nitrilases. Curr Opin Chem Biol 14:130–137. doi:10.1016/j.cbpa.2009.11.018
Meth-Cohn O, Wang M-X (1997) Rationalisation of the regioselective hydrolysis of aliphatic dinitriles with Rhodococcus rhodochrous AJ270. Chem Commun:1041–1042. doi: 10.1039/a700859g
Mukherjee C, Zhu D, Biehl ER, Hua L (2006) Exploring the synthetic applicability of a cyanobacterium nitrilase as catalyst for nitrile hydrolysis. Eur J Org Chem:5238–5242. doi: 10.1002/ejoc.200600699
Nagasawa T, Mauger J, Yamada H (1990) A novel nitrilase, arylacetonitrilase, of Alcaligenes faecalis JM3- purification and characterization. Eur J Biochem 194:765–772. doi:10.1111/j.1432-1033.1990.tb19467.x
Ni K, Wang H, Zhao L, Zhang M, Zhang S, Ren Y, Wei D (2013) Efficient production or R-(-)-mandelic acid in biphasic system by immobilized recombinant E. coli. J Biotechnol 167:433–440. doi:10.1016/j.jbiotec.2013.07.024
Petříčková A, Veselá AB, Kaplan O, Kubáč D, Uhnáková B, Malandra A, Felsberg J, Rinágelová A, Weyrauch P, Křen V, Bezouška K, Martínková L (2012) Purification and characterization of heterologously expressed nitrilases from filamentous fungi. Appl Microbiol Biotechnol 93:1553–1561. doi:10.1007/s00253-011-3525-7
Piotrowski M, Schönfelder S, Weiler EW (2001) The Arabidopsis thaliana isogene NIT4 and its orthologs in tobacco encode β-cyano-L-alanine hydratase/nitrilase. J Biol Chem 276:2616–2621. doi:10.1074/jbc.M007890200
Pouchert C, Behnke J, Sigma-Aldrich (1993) The Aldrich Library of 13C and 1H FT NMR Spectra. Chemical Co., Aldrich
Rey P, Rossi J-C, Taillades J, Gros G, Nore O (2004) Hydrolysis of nitriles using an immobilized nitrilase: applications to the synthesis of methionine hydroxy analogue derivatives. J Agric Food Chem 52:8155–8162. doi:10.1021/jf048827q
Rinágelová A, Kaplan O, Veselá AB, Chmátal M, Křenková A, Plíhal O, Pasquarelli F, Cantarella M, Martínková L (2014) Cyanide hydratase from Aspergillus niger K10: overproduction in Escherichia coli, purification, characterization and use in continuous cyanide degradation. Proc Biochem 49:445–450. doi:10.1016/j.procbio.2013.12.008
Robertson DE, Chaplin JA, DeSantis G, Podar M, Madden M, Chi E, Richardson T, Milan A, Miller M, Weiner DP, Wong K, McQuaid J, Farwell B, Preston LA, Tan X, Snead MA, Keller M, Mathur E, Kretz LP, Burk MJ, Short JM (2004) Exploring nitrilase sequence space for enantioselective catalysis. Appl Environ Microbiol 70:2429–2436. doi:10.1128/AEM.70.4.2429-2436.2004
Seffernick JL, Samanta SK, Louie TM, Wackett LP, Subramanian M (2009) Investigative mining of sequence data for novel enzymes: a case study with nitrilases. J Biotechnol 143:17–26. doi:10.1016/j.jbiotec.2009.06.004
Thuku RN, Brady D, Benedik MJ, Sewell BT (2009) Microbial nitrilases: versatile, spiral forming, industrial enzymes. J Appl Microbiol 106:703–727. doi:10.1111/j.1365-2672.2008.03941.x
Vejvoda V, Šveda O, Kaplan O, Přikrylová V, Elišáková V, Himl M, Kubáč D, Pelantová H, Kuzma M, Křen V, Martínková L (2007) Biotransformation of heterocyclic dinitriles by Rhodococcus erythropolis and fungal nitrilases. Biotechnol Lett 29:1119–1124. doi:10.1007/s10529-007-9364-z
Vergne-Vaxelaire C, Bordier F, Fossey A, Besnard-Gonnet M, Debard A, Mariage A, Pellouin V, Perret A, Petit J-L, Stam M, Salanoubat M, Weissenbach J, De Berardinis V, Zaparucha A (2013) Nitrilase activity screening on structurally diverse substrates: providing biocatalytic tools for organic synthesis. Adv Synth Catal 355:1763–1779. doi:10.1002/adsc.201201098
Veselá AB, Petříčková A, Weyrauch P, Martínková L (2013) Heterologous expression, purification and characterization of arylacetonitrilases from Nectria haematococca and Arthroderma benhamiae. Biocatal Biotransform 31:49–56. doi:10.3109/10242422.2012.758117
Veselá AB, Křenková A, Martínková L (2015) Exploring the potential of fungal arylacetonitrilases in mandelic acid synthesis. Mol Biotechnol 57:466–474. doi:10.1007/s12033-015-9840-y
Vorwerk S, Biernacki S, Hillebrand H, Janzik I, Müller A, Weiler EW, Piotrowski M (2001) Enzymatic characterization of the recombinant Arabidopsis thaliana nitrilase subfamily encoded by the NIT2/NIT1/NIT3-gene cluster. Planta 212:508–516. doi:10.1007/s004250000420
Wang H, Sun H, Wei D (2013) Discovery and characterization of a highly efficient enantioselective mandelonitrile hydrolase from Burkholderia cenocepacia J2315 by phylogeny-based enzymatic substrate specificity prediction. BMC Biotechnol 13:no. 14. doi:10.1186/1472-6750-13-14
Zhang, Z-J, Pan J., Liu J-F, Xu, J-H, He Y-C, Liu, Y-Y (2011) Significant enhancement of (R)-mandelic acid production by relieving substrate inhibition of recombinant nitrilase in toluene–water biphasic system. J Biotechnol 152:24–29. doi:10.1016/j.jbiotec.2011.01.013
Zhu D, Mukherjee C, Biehl ER, Hua L (2007a) Discovery of a mandelonitrile hydrolase from Bradyrhizobium japonicum USDA110 by rational genome mining. J Biotechnol 129:645–650. doi:10.1016/j.jbiotec.2007.02.001
Zhu D, Mukherjee C, Biehl ER, Hua L (2007b) Nitrilase-catalyzed selective hydrolysis of dinitriles and green access to the cyanocarboxylic acids of pharmaceutical importance. Adv Synth Catal 349:1667–1670. doi:10.1002/adsc.200700067
Zhu D, Mukherjee C, Yang Y, Rios BE, Gallagher DT, Smith NN, Biehl ER, Hua L (2008) A new nitrilase from Bradyrhizobium japonicum USDA 110. Gene cloning, biochemical characterization and substrate specificity. J Biotechnol 133:327–333. doi:10.1016/j.jbiotec.2007.10.001
Funding
This study was funded by the Czech Science Foundation (No. P504/11/034), the Technology Agency of the Czech Republic (grant no. TA01021368), and the Institute of Microbiology of the Czech Academy of Sciences (No. RVO61388971).
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Author information
Authors and Affiliations
Corresponding author
Additional information
Alicja B. Veselá and Lenka Rucká contributed equally to this work.
Electronic Supplementary material
ESM 1
(PDF 279 kb)
Rights and permissions
About this article
Cite this article
Veselá, A.B., Rucká, L., Kaplan, O. et al. Bringing nitrilase sequences from databases to life: the search for novel substrate specificities with a focus on dinitriles. Appl Microbiol Biotechnol 100, 2193–2202 (2016). https://doi.org/10.1007/s00253-015-7023-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-015-7023-1