Abstract
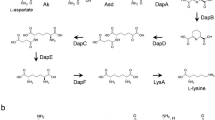
Streptomyces albulus NBRC14147 produces ɛ-poly-l-lysine (ɛ-PL), which is an amino acid homopolymer antibiotic. Despite the commercial importance of ɛ-PL, limited information is available regarding its biosynthesis; the l-lysine molecule is directly utilized for ɛ-PL biosynthesis. In most bacteria, l-lysine is biosynthesized by an aspartate pathway. Aspartokinase (Ask), which is the first enzyme in this pathway, is subject to complex regulation such as through feedback inhibition by the end-product amino acids such as l-lysine and/or l-threonine. S. albulus NBRC14147 can produce a large amount of ɛ-PL (1–3 g/l). We therefore suspected that Ask(s) of S. albulus could be resistant to feedback inhibition to provide sufficient l-lysine for ɛ-PL biosynthesis. To address this hypothesis, in this study, we cloned the ask gene from S. albulus and investigated the feedback inhibition of its gene product. As predicted, we revealed the feedback resistance of the Ask; more than 20% relative activity of Ask was detected in the assay mixture even with extremely high concentrations of l-lysine and l-threonine (100 mM each). We further constructed a mutated ask gene for which the gene product Ask (M68V) is almost fully resistant to feedback inhibition. The homologous expression of Ask (M68V) further demonstrated the increase in ɛ-PL productivity.





Similar content being viewed by others
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Bibb MJ, Findlay PR, Johnson MW (1984) The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene 30:157–166
Cuadrado Y, Fernandez M, Recio E, Aparicio JF, Martin JF (2004) Characterization of the ask–asd operon in aminoethoxyvinylglycine-producing Streptomyces sp. NRRL 5331. Appl Microbiol Biotechnol 64:228–236
Follettie MT, Peoples OP, Agoropoulou C, Sinskey AJ (1993) Gene structure and expression of the Corynebacterium flavum N13 ask–asd operon. J Bacteriol 175:4096–4103
Hamano Y, Nicchu I, Hoshino Y, Kawai T, Nakamori S, Takagi H (2005) Development of gene delivery systems for the ɛ-poly-l-lysine producer, Streptomyces albulus. J Biosci Bioeng 99:636–641
Hamano Y, Yoshida T, Kito M, Nakamori S, Nagasawa T, Takagi H (2006) Biological function of the pld gene product that degrades å-poly-l-lysine in Streptomyces albulus. Appl Microbiol Biotechnol 72:173–181
Hernando-Rico V, Martin JF, Santamarta I, Liras P (2001) Structure of the ask–asd operon and formation of aspartokinase subunits in the cephamycin producer ‘Amycolatopsis lactamdurans’. Microbiology 147:1547–1555
Hitchcock MJ, Hodgson B, Linforth JL (1980) Regulation of lysine- and lysine-plus-threonine-inhibitable aspartokinases in Bacillus brevis. J Bacteriol 142:424–432
Itzhaki RF (1972) Colorimetric method for estimating polylysine and polyarginine. Anal Biochem 50:569–574
Kahar P, Iwata T, Hiraki J, Park EY, Okabe M (2001) Enhancement of ɛ-polylysine production by Streptomyces albulus strain 410 using pH control. J Biosci Bioeng 91:190–194
Kalinowski J, Cremer J, Bachmann B, Eggeling L, Sahm H, Puhler A (1991) Genetic and biochemical analysis of the aspartokinase from Corynebacterium glutamicum. Mol Microbiol 5:1197–1204
Kawai T, Kubota T, Hiraki J, Izumi Y (2003) Biosynthesis of ɛ-poly-l-lysine in a cell-free system of Streptomyces albulus. Biochem Biophys Res Commun 311:635–640
Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA (2000) Practical Streptomyces Genetics. The John Innes Foundation, Norwich, UK
Kobashi N, Nishiyama M, Yamane H (2001) Characterization of aspartate kinase III of Bacillus subtilis. Biosci Biotechnol Biochem 65:1391–1394
Ogawa-Miyata Y, Kojima H, Sano K (2001) Mutation analysis of the feedback inhibition site of aspartokinase III of Escherichia coli K-12 and its use in l-threonine production. Biosci Biotechnol Biochem 65:1149–1154
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
Shiio I (1982) Metabolic regulation and over-production of amino acids. In: Krumphanzl V, Sikyta B, Vanek Z (eds) Overproduction of microbial products. Academic, London, pp 463–472
Shiio I, Miyajima R (1969) Concerted inhibition and its reversal by end products of aspartate kinase in Brevibacterium flavum. J Biochem (Tokyo) 65:849–859
Shima S, Sakai H (1977) Polylysine produced by Streptomyces. Agric Biol Chem 41:1807–1809
Shima S, Sakai H (1981a) Poly-l-lysine produced by Streptomyces. III. Chemical studies. Agric Biol Chem 45:2503–2508
Shima S, Sakai H (1981b) Poly-l-lysine produced by Streptomyces. II. Taxonomy and fermentation studies. Agric Biol Chem 45:2497–2502
Shima S, Matsuoka H, Sakai H (1982) Inactivation of bacteriopharges by ɛ-poly-l-lysine produced by Streptomyces. Agric Biol Chem 46:1917–1919
Shima S, Oshima S, Sakai H (1983) Biosynthesis of ɛ-poly-l-lysine by washed mycelium of Streptomyces albulus No. 346. Nippon Nogei KagakuKaishi 57:221–226
Shima S, Matsuoka H, Iwamoto T, Sakai H (1984) Antimicrobial action of ɛ-poly-l-lysine. J Antibiot (Tokyo) 37:1449–1455
Theze J, Margarita D, Cohen GN, Borne F, Patte JC (1974) Map** of the structural genes of the three aspartokinases and of the two homoserine dehydrogenases of Escherichia coli K-12. J Bacteriol 117:133–143
Tunca S, Yilmaz EI, Piret J, Liras P, Ozcengiz G (2004) Cloning, characterization and heterologous expression of the aspartokinase and aspartate semialdehyde dehydrogenase genes of cephamycin C-producer Streptomyces clavuligerus. Res Microbiol 155:525–534
Zhang JJ, Paulus H (1990) Desensitization of Bacillus subtilis aspartokinase I to allosteric inhibition by meso-diaminopimelate allows aspartokinase I to function in amino acid biosynthesis during exponential growth. J Bacteriol 172:4690–4693
Zhang JJ, Hu FM, Chen NY, Paulus H (1990) Comparison of the three aspartokinase isozymes in Bacillus subtilis Marburg and 168. J Bacteriol 172:701–708
Zhang W, Jiang W, Zhao G, Yang Y, Chiao J (1999) Sequence analysis and expression of the aspartokinase and aspartate semialdehyde dehydrogenase operon from rifamycin SV-producing Amycolatopsis mediterranei. Gene 237:413–419
Zhang WW, Jiang WH, Zhao GP, Yang YL, Chiao JS (2000) Expression in Escherichia coli, purification and kinetic analysis of the aspartokinase and aspartate semialdehyde dehydrogenase from the rifamycin SV-producing Amycolatopsis mediterranei U32. Appl Microbiol Biotechnol 54:52–58
Acknowledgments
This work was supported in part by a grant from Chisso Corporation.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Hamano, Y., Nicchu, I., Shimizu, T. et al. ɛ-Poly-l-lysine producer, Streptomyces albulus, has feedback-inhibition resistant aspartokinase. Appl Microbiol Biotechnol 76, 873–882 (2007). https://doi.org/10.1007/s00253-007-1052-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-007-1052-3