Abstract
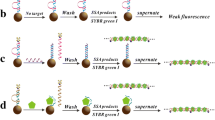
Dye-encapsulating liposomes can serve as signaling reagents in biosensors and biochemical assays in place of enzymes or fluorophores. Detailed here is the use and preparation of streptavidin-coupled liposomes which offer a universal approach to biotinylated target detection. The universal approach provides two advantages, i.e. only one type of liposome is necessary despite varying target and probe sequences and the hybridization event can take place in the absence of potential steric hindrance occurring from liposomes directly conjugated to probes. One objective of this work was to optimize the one-step conjugation of SRB-encapsulating liposomes to streptavidin using EDC. Liposome, EDC, streptavidin concentrations, and reaction times were varied. The optimal coupling conditions were found to be an EDC:carboxylated lipid:streptavidin molar ratio of 600:120:1 and a reaction time of 15 min. The second goal was to utilize these liposomes in sandwich hybridization microtiter plate-based assays using biotinylated reported probes as biorecognition elements. The assay was optimized in terms of probe spacer length, probe concentration, liposome concentration, and streptavidin coverage. Subsequently, the optimized protocol was applied to the detection of DNA and RNA sequences. A detection limit of 1.7 pmol L−1 and an assay range spanning four orders of magnitude (5 pmol L−1−50 nmol L−1) with a coefficient of variation ≤5.8% was found for synthetic DNA. For synthetic RNA the LOQ was half that of synthetic DNA. A comparison was made to alkaline phosphatase-conjugated streptavidin for detection which yielded a limit of quantitation approximately 80 times higher than that for liposomes in the same system. Thus, liposomes and the optimized sandwich hybridization method are well suited for detecting single-stranded nucleic acid sequences and compares favorably to other sandwich hybridization schemes recently described in the literature. The assay was then used successfully for the clear detection of mRNA amplified by nucleic acid sequence-based amplification (NASBA) isolated from as little as one Cryptosporidium parvum oocyst. The detection of mRNA from oocysts isolated from various water sample types using immunomagnetic separation was also assessed. Finally, to prove the wider applicability and sensitivity of this universal method, RNA amplified from the atxA gene of Bacillus anthracis was detected when the input to the preceding NASBA reaction was as low as 1.2 pg. This highly sensitive liposome-based microtiter plate assay is therefore a platform technology allowing for high throughput and wide availability for routine clinical and environmental laboratory applications.










Similar content being viewed by others
Abbreviations
- AP:
-
Alkaline phosphatase
- BSA:
-
Bovine serum albumin
- DIG:
-
Digoxin
- DPPC:
-
1,2-Dipalmitoyl-sn-glycero-3-phosphocholine
- DPPE:
-
1,2-Dipalmitoyl-sn-glycero-3-phosphoethanolamine
- DPPG:
-
1,2-Dipalmitoyl-sn-glycero-3-[phospho-rac-(1-glycerol)] sodium salt
- EDC:
-
1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide
- EDTA:
-
Ethylenediamine tetraacetic acid
- HEPES:
-
N-2-Hydroxyethylpiperazine-N′-2-ethanesulfonic acid
- HPV:
-
Human papillomavirus
- HSS:
-
HEPES-saline-sucrose
- IMS:
-
Immunomagnetic separation
- IPTG:
-
Isopropyl β-d-1-thiogalactopyranoside
- LNA:
-
Locked nucleic acid
- MES:
-
2-(4-Morpholino)ethanesulfonic acid
- NASBA:
-
Nucleic acid-based sequence amplification
- LOQ:
-
Limit of quantitation
- OG:
-
n-Octyl-β-d-glucopyranoside
- PBS:
-
Phosphate-buffered saline
- PCR:
-
Polymerase chain reaction
- SRB:
-
Sulforhodamine B
- StAv:
-
Streptavidin
- TBS:
-
Tris buffered saline
References
Marx J (2000) Science 289:1670–1672
Ranki M, Virtanen M, Palva A, Laaksonen M, Pettersson R, Kaariainen L, Halonen P, Soderlund H (1983) Curr Top Microbiol Immunol 104:307–318
Parkkinen S, Mantyjarvi R, Syrjanen K, Ranki M (1986) J Med Virol 20:279–288
Lee CY, Panicker G, Bej AK (2003) J Microbiol Methods 53:199–209
Namimatsu T, Tsuna M, Imai Y, Futo S, Mitsuse S, Sakano T, Sato S (2000) J Vet Med Sci 62:615–619
Palkovics L, Burgyan J, Balazs E (1994) Res Virol 145:387–392
Baeumner AJ, Leonard B, McElwee J, Montagna RA (2004) Anal Bioanal Chem 380:15–23
Ranki M, Palva A, Virtanen M, Laaksonen M, Soderlund H (1983) Gene 21:77–85
Leskela T, Tilsala-Timisjarvi A, Kusnetsov J, Neubauer P, Breitenstein A (2005) J Microbiol Methods 62:167–179
Zhang N, Appella DH (2007) J Am Chem Soc 129:8424–8425
Edwards KA, Baeumner AJ (2006) Anal Chem 78:1958–1966
Ishii JK, Ghosh SS (1993) Bioconjug Chem 4:34–41
Casademont I, Bizet C, Chevrier D, Guesdon JL (2000) Mol Cell Probes 14:233–240
Aubin JT, Boulay D, Chapus A, Brechot C, Laure F, Agut H (1995) Res Virol 146:75–79
Li ZP, Liu CH, Fan YS, Duan XR (2007) Anal Bioanal Chem 387:613–618
Rule GS, Montagna RA, Durst RA (1996) Clin Chem 42:1206–1209
Edwards KA, Baeumner AJ (2006) Anal Bioanal Chem 386:1335–1343
Edwards KA, Baeumner AJ (2006) Talanta 68:1421–1431
Rongen HAH, Bult A, vanBennekom WP (1997) J Immunol Methods 204:105–133
Locascio LE, Hong JS, Gaitan M (2002) Electrophoresis 23:799–804
Park JW, Kirpotin DB, Hong K, Shalaby R, Shao Y, Nielsen UB, Marks JD, Papahadjopoulos D, Benz CC (2001) J Controlled Release 74:95–113
Pardridge WM (2007) Adv Drug Deliv Rev
Edwards KA, Baeumner AJ (2007) Anal Chem 79:1806–1815
Campbell CH, Miller AL, Rome LH (1983) Biochem J 214:413–419
Madhankumar AB, Slagle-Webb B, Mintz A, Sheehan JM, Connor JR (2006) Mol Cancer Ther 5:3162–3169
Willis MC, Collins BD, Zhang T, Green LS, Sebesta DP, Bell C, Kellogg E, Gill SC, Magallanez A, Knauer S, Bendele RA, Gill PS, Janjic N (1998) Bioconjug Chem 9:573–582
Edwards KA, March JC (2007) Anal Biochem 368:39–48
Ahn-Yoon S, DeCory TR, Baeumner AJ, Durst RA (2003) Anal Chem 75:2256–2261
Ahn-Yoon S, DeCory TR, Durst RA (2004) Anal Bioanal Chem 378:68–75
Hutchinson FJ, Jones MN (1988) FEBS Lett 234:493–496
Liautard JP, Vidal M, Philippot JR (1985) Cell Biol Int Rep 9:1123–1137
Duzgunes N, Pretzer E, Simoes S, Slepushkin V, Konopka K, Flasher D, de Lima MC (1999) Mol Membr Biol 16:111–118
Chen C-S, Baeumner A, Durst R (2005) Talanta 67:205–211
Plant AL, Brizgys MV, Locasio-Brown L, Durst RA (1989) Anal Biochem 176:420–426
Baeumner AJ, Jones C, Wong CY, Price A (2004) Anal Bioanal Chem 378:1587–1593
Wen HW, DeCory TR, Borejsza-Wysocki W, Durst RA (2006) Talanta 68:1264–1272
Weber PC, Ohlendorf DH, Wendoloski JJ, Salemme FR (1989) Science 243:85–88
Martin FJ, Papahadjopoulos D (1982) J Biol Chem 257:286–288
Martin FJ, Hubbell WL, Papahadjopoulos D (1981) Biochemistry 20:4229–4238
Weissig V, Lasch J, Klibanov AL, Torchilin VP (1986) FEBS Lett 202:86–90
Bendas G, Vogel J, Bakowski U, Krause A, Muller J, Rothe U (1997) Biochim Biophys Acta 1325:297–308
Bredehorst R, Ligler FS, Kusterbeck AW, Chang EL, Gaber BP, Vogel CW (1986) Biochemistry 25:5693–5698
MacKenzie WR, Hoxie NJ, Proctor ME, Gradus MS, Blair KA, Peterson DE, Kazmierczak JJ, Addiss DG, Fox KR, Rose JB et al (1994) N Engl J Med 331:161–167
Sischo WM, Atwill ER, Lanyon LE, George J (2000) Prev Vet Med 43:253–267
Fayer R, Trout JM, Lewis EJ, Santin M, Zhou L, Lal AA, **ao L (2003) Parasitol Res 89:141–145
Nime FA, Burek JD, Page DL, Holscher MA, Yardley JH (1976) Gastroenterology 70:592–598
Dragon DC, Bader DE, Mitchell J, Woollen N (2005) Appl Environ Microbiol 71:1610–1615
Dahlgren CM, Buchanan LM, Decker HM, Freed SW, Phillips CR, Brachman PS (1960) Am J Hyg 72:24–31
Sanderson WT, Stoddard RR, Echt AS, Piacitelli CA, Kim D, Horan J, Davies MM, McCleery RE, Muller P, Schnorr TM, Ward EM, Hales TR (2004) J Appl Microbiol 96:1048–1056
Cook N (2003) J Microbiol Methods 53:165–174
Keer JT, Birch L (2003) J Microbiol Methods 53:175–183
Koppel DE (1972) J Chem Phys 57:4814
Frisken BJ (2001) Appl Optics 40:4087–4091
Connelly J, Nugen S, Borejsza-Wysocki W, Durst R, Montagna R, Baeumner A (2008) Anal Bioanal Chem
Bartlett GR (1959) J Biol Chem 234:466–468
Fiske CH, Subbarow Y (1925) J Biol Chem 66:375–400
Singh AK, Kilpatrick PK, Carbonell RG (1996) Biotechnol Prog 12:272–280
Ege C, Lee KY (2004) Biophys J 87:1732–1740
Israelachvili JN, Mitchell DJ (1975) Biochim Biophys Acta 389:13–19
Hartley HA, Baeumner AJ (2003) Anal Bioanal Chem 376:319–327
Boom R, Sol CJ, Salimans MM, Jansen CL, Wertheim-van Dillen PM, van der Noordaa J (1990) J Clin Microbiol 28:495–503
Gottschalk PG, Dunn JR (2005) Anal Biochem 343:54–65
Gottschalk PG, Dunn JR, Vol Tech note 3022 Bio-Rad Laboratories Inc, Hercules CA USA
Thompson V, Schatzlein D, Mercuro D (2003) Spectroscopy 18:112–114
Edwards KA, Baeumner AJ (2006) Anal Bioanal Chem 386:1613–1623
Grabarek Z, Gergely J (1990) Anal Biochem 185:131–135
Bogdanov AA Jr, Klibanov AL, Torchilin VP (1988) FEBS Lett 231:381–384
Kohsaka H, Taniguchi A, Richman DD, Carson DA (1993) Nucleic Acids Res 21:3469–3472
Running JA, Urdea MS (1990) Biotechniques 8:276–279
Nickerson DA, Kaiser R, Lappin S, Stewart J, Hood L, Landegren U (1990) Proc Natl Acad Sci USA 87:8923–8927
Jeltsch A, Fritz A, Alves J, Wolfes H, **oud A (1993) Anal Biochem 213:234–240
Yang B, Viscidi R, Yolken R (1993) Anal Biochem 213:422–425
Nikiforov TT, Rogers YH (1995) Anal Biochem 227:201–209
Nikiforov TT, Rendle RB, Goelet P, Rogers YH, Kotewicz ML, Anderson S, Trainor GL, Knapp MR (1994) Nucleic Acids Res 22:4167–4175
Nikiforov TT, Rendle RB, Kotewicz ML, Rogers YH (1994) PCR Methods Appl 3:285–291
Nunc (2007) Vol 2007 Nalge Nunc International
Dai Z, Sirard JC, Mock M, Koehler TM (1995) Mol Microbiol 16:1171–1181
Baeumner AJ, Humiston M, Montagna RA, Durst RA (2001) Anal Chem 73:1176–1180
Esch MB, Baeumner AJ, Durst RA (2001) Anal Chem 73:3162–3167
Rautio J, Barken KB, Lahdenpera J, Breitenstein A, Molin S, Neubauer P (2003) Microb Cell Fact 2:4
Chen Y, Chi Y, Wen H, Lu Z (2007) Anal Chem 79:960–965
Goral VN, Zaytseva NV, Baeumner AJ (2006) Lab Chip 6:414–421
Wicks B, Cook DB, Barer MR, O’Donnell AG, Self CH (1998) Anal Biochem 259:258–264
Nugen SR, Leonard B, Baeumner AJ (2007) Biosens Bioelectron 22:2442–2448
Clavel C, Masure M, Levert M, Putaud I, Mangeonjean C, Lorenzato M, Nazeyrollas P, Gabriel R, Quereux C, Birembaut P (2000) Diagn Mol Pathol 9:145–150
Neely LA, Patel S, Garver J, Gallo M, Hackett M, McLaughlin S, Nadel M, Harris J, Gullans S, Rooke J (2006) Nat Methods 3:41–46
Kauppinen S, Vester B, Wengel J (2006) Handb Exp Pharmacol:405–422
Acknowledgements
The authors are grateful to Ravi Sood for his preliminary investigations into optimizations of microtiter plate-based DNA hybridization methodology. John Connelly provided the C. parvum NASBA amplicons from environmental water samples for this work. This project was funded in part by the CD4 Initiative, Imperial College, London, UK.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Edwards, K.A., Curtis, K.L., Sailor, J.L. et al. Universal liposomes: preparation and usage for the detection of mRNA. Anal Bioanal Chem 391, 1689–1702 (2008). https://doi.org/10.1007/s00216-008-1992-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-008-1992-1