Abstract
SARS-CoV-2 is an enveloped positive-sense RNA virus, contain crown-like spikes on its surface, exceptional of large RNA genome, and a special replication machinery. Common symptoms of SARS-CoV-2 include cough, common cold, fever, sore throat, and a variety of severe acute respiratory disease (SARD) such as pneumonia. SARS-CoV-2 infects epithelial cells, T-cells, macrophages, and dendritic cells and also influences the production and implantation of pro-inflammatory cytokines and chemokines. Repurposing of various drugs during this emergency condition can reduce the rate of mortality as well as time and cost. Two druggable protein and enzyme targets have been selected in this review article due to their crucial role in the viral life cycle. The eukaryotic translation initiation factor (eIF4A), cyclophilin, nucleocapsid protein, spike protein, Angiotensin-converting enzyme 2 (ACE2), 3-chymotrypsin-like cysteine protease (3CLpro), and RNA-dependent RNA polymerase (RdRp) play significant role in early and late phase of SARS-CoV-2 replication and translation. This review paper is based on the rationale of inhibiting of various SARS-CoV-2 proteins and enzymes as novel therapeutic approaches for the management and treatment of patients with SARS-CoV-2 infection. We also discussed the structural and functional relationship of different proteins and enzymes to develop therapeutic approaches for novel coronavirus SARS-CoV-2.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
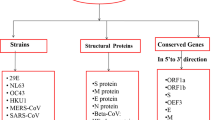
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) is a highly transmissible and pathogenic coronavirus that mainly affects the human respiratory system. SARS-CoV-2 is responsible for two distinct endemics like Middle East respiratory syndrome (MERS) and acute respiratory syndrome (SARS), which have significant affected on public health (Raoult et al. 2020). SARS-CoV-2 is named due to the presence of crown-like spikes on their surface and consisted of four sub-groups, called as alpha, beta, gamma, and delta (Fehr and Perlman, 2015). It is a positive sense-stranded RNA virus with 29,891 bases; among these, 96% bases are identical to a bat coronavirus (CoVs), at the full level of genome stage, and share 79.6% of gene similarity with SARS-CoV (Denison et al. 2011). SARS-CoV-2 encodes spike (S) protein consisting of a receptor-binding domain (RBD) that binds to the angiotensin-converting enzyme-2 (ACE-2) of humans and facilitates membrane fusion as well as virus uptake into human lungs (Fig. 1) (Hofmann and Pöhlmann, 2004). SARS-CoV-2 enter into human cells and capture the protein synthesis machinery to synthesize the viral proteins for replication and proliferation (Hofmann and Pöhlmann, 2004). SARS-CoV-2 contains the largest genomic structure (26.4–31.7 kb) among all known RNA viruses. Large numbers of small open reading frames (ORFs) are present between the various conserved genes [ORF1ab, spike (S), envelope (E), membrane (M), nucleocapsid (N)] and the nucleocapsid genes of various CoVs lineages (Mousavizadeh and Ghasemi, 2020). The viral genomes consist of distinctive characteristics, including a unique N-terminal fragment within the spike protein. Genes for main structural proteins in all SARS-CoV-2 occur in 5′–3′ order, such as S, E, M, and N. A typical SARS-CoV-2 contains at least six ORFs in their genome. ORF1a and ORF1b provide a frameshift between two polypeptides that are pp1a and pp1ab (Prajapat et al. 2020). These polypeptides are converted into 16 nsps (nsp1-16) by virally encoded chymotrypsin-like protease (3CLpro) or main protease (Mpro) and one or two papain-like proteases. ORFs 10,11 encode four specific structural proteins containing S, E, M, N proteins on one-third of the genome near to the 3′-terminus (van Boheemen et al. 2012). In addition to these four main structural proteins, such as HE protein, 3a/b protein and 4a/b protein are encoded by various CoVs (Fig. 2) (Chen et al. 2020). Such mature proteins are responsible for maintaining genomic structural integrity maintenance and virus replication roles.
The genomic structure and phylogenetic of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2): a The phylogenetic tree of coronavirus with the new COVID-19 shown in green color. b The genome structure of four genera of coronaviruses (CoVs): two long polypeptides with 16 nonstructural proteins initiated from Pp1a to pp1b represent. E, S, M, and N are consisted of the four structural proteins envelope, spike, membrane, and nucleocapsid. Abbreviations: CoVs, coronavirus; HE, hemagglutinin-esterase. HCoV, human coronavirus; HKU, coronaviruses identified by Hong Kong University; MHV, murine hepatitis virus; IBV, infectious bronchitis virus; TGEV, transmissible gastroenteritis virus; HCoV-229E, human coronavirus OC43; MERS‐CoV, Middle East respiratory syndrome coronavirus
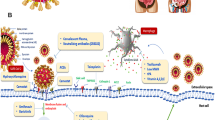
The genome gets transcribed after the virus enters into host cell. The reproduction and transcription of the CoVs genome occur on cytoplasmic membrane and regulate by the viral replicate (Shulla et al. 2011). It is assumed that the replicase complex has consisted of approximately 16 subunits and a various cellular protein. In addition to RNA-dependent RNA polymerase (RdRp), RNA helicase, and activities of proteases which are common in many RNA viruses, CoVs replicase is known to use a variety of RNA-dependent processing enzymes which are not present in other RNA viruses, including a putative specific sequence of endoribonuclease, 3′- to 5′-exoribonuclease, 2′-O-ribose methyltransferase, ADP ribose 1′-phosphatase, and cyclic phosphodiesterase behaviors in a subset of group 2 CoVs (Sola et al. 2015; Ziebuhr, 2005). The proteins are packaged on the cellular membranes and genomic RNA is introduced by budding from the internal cell membrane as the mature particles emerge (Almazán et al. 2006). SARS-CoV-2 N-proteins have 3 distinct and highly conserved domains include 2 structural and independently folded structural regions, known as N terminal domain (NTD/domain 1) and C-terminal domain (CTD/domain 3), separated by intrinsic disordered central region (RNA-binding domain/domain 2) (Fig. 3) (Huang et al. 2004).
Structure of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) nucleocapsid protein and target sites of potential antiviral agents. The virion enters by endocytosis or direct fusion of cell through viral membranes. The viral genome is translated into two polyproteins, which are cleaved by two viral proteases (3CLpro PLpro) to generate a large replication and transcription complex orchestrating genome replication and synthesis of mRNAs. New viral genomes recruit viral structural proteins to generate new virions released by exocytosis process. Red arrow indicates the potential inhibitors used to inhibit various targets. Abbreviations: 3CLpro, chymotrypsin-like protease; PLpro, papain-like protease; 3UTR, 3 untranslated region; 5UTR, 5 untranslated region; pp 1 ab, polypeptide 1ab; CYP, cyclophilin; RdRP, RNA-dependent RNA polymerase
Number of patients were hospitalized with initial diagnosis of unknown pneumonia in December 2019. Available studies have indicated that bat may be the potential reservoir of SARS-CoV, which cause serious illness in humans and agricultural animals. However, there is no confirmation to date that SARS-CoV-2 was originated from the seafood market but bats are the ideal repository for a variety of SARS-CoV-2, including MERS-CoV and SARS-CoV (Guo et al., 2020). The genome sequencing of COVID-19 was analyzed and found 96.2% similar to Bat CoV RaTG13 because both types of viruses might be shared the same ancestor (Zhang et al. 2020). The exact in vivo effect of these drugs is yet unclear, however, and further finding may confirm the mechanism of inhibiting SARS‐CoV‐2 and reducing associated infections.
Neuraminidase and M2 ion-channel protein
Neuraminidase plays an important role in cleavage of terminal sialic acid residues from glycoconjugates and is essential for virus replication and infectivity (Akhtar, 2020). Neuraminidase inhibitors (oseltamivir, zanamivir, and peramivir) are not expected to be effective against COVID-19 due to absence of this enzyme in SARS-CoV-2. Moreover, oseltamivir with ganciclovir and lopinavir/ritonavir was found beneficial to treat COVID-19 infections in Wuhan city (Chu et al. 2020; Huang et al. 2020). In silico study also found that combination of oseltamivir-lopinavir-ritonavir c had synergistic effects against SARS-CoV-2 (Muralidharan et al. 2020). In Indonesia and Singapore, oseltamivir is currently being used as a recommended COVID-19 treatment option.
The M2 channel protein is essential viral envelope protein for maintaining pH across the viral envelope, and plays an important role during entry and movement across the trans-Golgi host cell membrane during viral maturation (Skehel et al. 1978). Previous studies have shown that amantadine could block the p7 protein of HCV, which is crucial to form ion channels in host cell membranes (Griffin et al. 2003). In 1973, amantadine was found to have a potent antiviral effect against coronavirus 229E in vitro, and later, it was able to block SARS-CoV’s protein-membrane channel activity. Furthermore, amantadine showed good antiviral activity against SARS-CoV-2 (Frediansyah et al. 2020) but more molecular analysis determines its specificity toward particular statin.
Conclusion and future perspective
SARS-CoV-2 a is single-stranded positive RNA virus and uses several host viral proteins and cellular components to complete its replication cycle, including the steps of viral entry, replication. Development of drug and vaccine against the SARS-CoV-2 is a challenging job due to lack of predictive in vitro and animal model, insufficient knowledge regarding underlying mechanism of action of disease, lack of targets and biomarkers, and a high rate of failed clinical trials. We need to know more structural biology, life cycle details, which can speed up the drug/vaccine development process against SARS-CoV-2. Again, to avoid these types of pandemic insult, strict vigilance of viral infection and understanding of viral protein and enzyme structure are necessary. Several series of small-molecule SARS-CoV-2 inhibitors targeting these protein and enzymes (eIF4A, cyclophilin, nucleocapsid protein, spike protein, ACE2, 3CLpro, and RdRp) have discussed in our article. However, most of them were tested in vitro, while only a small percentage of these compounds have been evaluated in animal study, and few have advanced into clinical trial study. Therefore, further studies should be focused on exploring novel strategies to identify new anti-CoVs compounds, elaborated their mechanism of action, improving the efficacy of anti-CoVs compounds, and evaluating the in vivo efficacy and safety of these compounds in different preclinical and clinical studies. Furthermore, development of small-molecule CoVs inhibitors with high efficacy and low toxicity will be brought for treatment of SARS-CoV-2 infection and related disease in the future.
Data availability
Not applicable.
Abbreviations
- SARS-CoV-2:
-
Severe acute respiratory syndrome coronavirus-2
- SARD:
-
Severe acute respiratory disease
- BCoV:
-
Bovine CoVs infectious
- IBV:
-
Bronchitis virus
- TGEV:
-
Transmissible Gastric Enteritis Virus
- RBD:
-
Receptor Binding Domain
- DPP4:
-
Dipeptidyl-peptidase 4
- eIF4A:
-
Eukaryotic translation initiation factor 4 A
- Cyps:
-
Cyclophilins
- ALV:
-
Alisporivir
- HCV:
-
Hepatitis C virus
- NTD:
-
N terminal domain
- CTD:
-
C-terminal domain
- IRF-3:
-
Interferon regulatory factor-3
- ACE2:
-
Angiotensin-converting enzyme
- RdRp:
-
RNA-dependent RNA polymerase
References
Abd El-Aziz TM, Stockand JD (2020) Recent progress and challenges in drug development against COVID-19 coronavirus (SARS-CoV-2)-an update on the status. Infect Genet Evol 83:104327–104337
Adedeji AO, Severson W, Jonsson C, Singh K, Weiss SR, Sarafianos SG (2013) Novel inhibitors of severe acute respiratory syndrome coronavirus entry that act by three distinct mechanisms. J Virol 87(14):8017–8028. https://doi.org/10.1128/jvi.00998-13
Akhtar MJ (2020) COVID19 inhibitors: prospective therapeutics. Bioorg Chem 101:104027. https://doi.org/10.1016/j.bioorg.2020.104027
Almazán F, DeDiego ML, Galán C, Escors D, Álvarez E, Ortego J, Sola I, Zuñiga S, Alonso S, Moreno JL, Nogales A (2006) Construction of a severe acute respiratory syndrome coronavirus infectious cDNA clone and a replicon to study coronavirus RNA synthesis. J Virol 80(21):10900–10906
Al-Tawfiq JA, Al-Homoud AH, Memish ZA (2020) Remdesivir as a possible therapeutic option for the COVID-19. Travel Med Infect Dis 34:101615–101617. https://doi.org/10.1016/j.tmaid.2020.101615
Amanat F, White KM, Miorin L, Strohmeier S, McMahon M, Meade P, Liu WC, Albrecht RA, Simon V, Martinez-Sobrido L, Moran T (2020) An in vitro microneutralization assay for SARS-CoV-2 serology and drug screening. Curr Protoc Microbiol 58(1):e108. https://doi.org/10.1002/cpmc.108
Andersen PI, Ianevski A, Lysvand H, Vitkauskiene A, Oksenych V, Bjørås M, Telling K, Lutsar I, Dampis U, Irie Y, Tenson T (2020) Discovery and development of safe-in-man broad-spectrum antiviral agents. Int J Infect Dis 93:268–276. https://doi.org/10.1016/j.ijid.2020.02.018
Andreou AZ, Harms U, Klostermeier D (2017) eIF4B stimulates eIF4A ATPase and unwinding activities by direct interaction through its 7-repeats region. RNA Biol 14(1):113–123. https://doi.org/10.1080/15476286.2016.1259782
Andreou AZ, Klostermeier D (2013) The DEAD-box helicase eIF4A: paradigm or the odd oneout? RNA Biol 10(1):19–32. https://doi.org/10.4161/rna.21966
Astuti I (2020) Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): an overview of viral structure and host response. Diabetes Metabolic Syndrome: Clinical Research & Reviews 14(4):407–412. https://doi.org/10.1016/j.dsx.2020.04.020
Báez-Santos YM, John SE, Mesecar AD (2015) The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds. Antiviral Res 115:21–38. https://doi.org/10.1016/j.antiviral.2014.12.015
Biedenkopf N, Lange-Grünweller K, Schulte FW, Weißer A, Müller C, Becker D, Becker S, Hartmann RK, Grünweller A (2017) The natural compound silvestrol is a potent inhibitor of Ebola virus replication. Antiviral Res 137:76–81. https://doi.org/10.1016/j.antiviral.2016.11.011
Cannalire R, Stefanelli I, Cerchia C, Beccari AR, Pelliccia S, Summa V (2020) SARS-CoV-2 entry inhibitors: small molecules and peptides targeting virus or host cells. Int J Mol Sci 21(16):5707. https://doi.org/10.3390/ijms21165707
Cao YC, Deng QX, Dai SX (2020) Remdesivir for severe acute respiratory syndrome coronavirus 2 causing COVID-19: an evaluation of the evidence. Travel Med Infect Dis 35:101647–101652. https://doi.org/10.1016/j.tmaid.2020.101647
Carbajo-Lozoya J, Müller MA, Kallies S, Thiel V, Drosten C, von Brunn A (2012) Replication of human coronaviruses SARS-CoV, HCoV-NL63 and HCoV-229E is inhibited by the drug FK506. Virus Res 165(1):112–117. https://doi.org/10.1016/j.virusres.2012.02.002
Chang CK, Hou MH, Chang CF, Hsiao CD, Huang TH (2014) The SARS coronavirus nucleocapsid protein–forms and functions. Antiviral Res 103:39–50. https://doi.org/10.1016/j.antiviral.2013.12.009
Chen Y, Liu Q, Guo D (2020) Emerging coronaviruses: genome structure, replication, and pathogenesis. J Med Virol 92(4):418–423. https://doi.org/10.1002/jmv.25681
Chowdhury P (2020) In silico investigation of phytoconstituents from Indian medicinal herb ‘Tinospora cordifolia (giloy)’against SARS-CoV-2 (COVID-19) by molecular dynamics approach. J Biomol Struct Dynam 1-8. https://doi.org/10.1080/07391102.2020.1803968
Chu DK, Pan Y, Cheng SM, Hui KP, Krishnan P, Liu Y, Ng DY, Wan CK, Yang P, Wang Q, Peiris M (2020) Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin Chem 66(4):549–555. https://doi.org/10.1093/clinchem/hvaa029
Chuck CP, Chong LT, Chen C, Chow HF, Wan DC, Wong KB (2010) Profiling of substrate specificity of SARS-CoV 3CLpro. PLoS ONE 5(10):13197. https://doi.org/10.1371/journal.pone.0013197
Coutard B, Valle C, de Lamballerie X, Canard B, Seidah NG, Decroly E (2020) The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res 176:104742. https://doi.org/10.1016/j.antiviral.2020.104742
Dawar FU, Tu J, Khattak MN, Mei J, Lin L (2017) Cyclophilin A: a key factor in virus replication and potential target for anti-viral therapy. Curr Issues Mol Biol 21:1–20. https://doi.org/10.21775/cimb.021.001
De Wilde AH, Falzarano D, Zevenhoven-Dobbe JC, Beugeling C, Fett C, Martellaro C, Posthuma CC, Feldmann H, Perlman S, Snijder EJ (2017) Alisporivir inhibits MERS-and SARS-coronavirus replication in cell culture, but not SARS-coronavirus infection in a mouse model. Virus Res 228:7–13. https://doi.org/10.1016/j.virusres.2016.11.011
DeDiego ML, Nieto-Torres JL, Regla-Nava JA, Jimenez-Guardeño JM, Fernandez-Delgado R, Fett C, Castaño-Rodriguez C, Perlman S, Enjuanes L (2014) Inhibition of NF-κB-mediated inflammation in severe acute respiratory syndrome coronavirus-infected mice increases survival. J Virol 88(2):913–924. https://doi.org/10.1128/JVI.02576-13
Denison MR, Graham RL, Donaldson EF, Eckerle LD, Baric RS (2011) Coronaviruses: an RNA proofreading machine regulates replication fidelity and diversity. RNA Biol 8(2):270–279. https://doi.org/10.4161/rna.8.2.15013
Dmitriev SE, Vladimirov DO, Lashkevich KA (2020) A quick guide to small-molecule inhibitors of eukaryotic protein synthesis. Biochem Mosc 85(11):1389–1421. https://doi.org/10.1134/s0006297920110097
Du L, He Y, Zhou Y, Liu S, Zheng BJ, Jiang S (2009) The spike protein of SARS-CoV—a target for vaccine and therapeutic development. Nat Rev Microbiol 7(3):226–236. https://doi.org/10.1038/nrmicro2090
Eslami G, Mousaviasl S, Radmanesh E, Jelvay S, Bitaraf S, Simmons B, Wentzel H, Hill A, Sadeghi A, Freeman J, Salmanzadeh S (2020) The impact of sofosbuvir/daclatasvir or ribavirin in patients with severe COVID-19. J Antimicrob Chemother 75(11):3366–3372. https://doi.org/10.1093/jac/dkaa331
Ewart GD, Mills K, Cox GB, Gage PW (2002) Amiloride derivatives block ion channel activity and enhancement of virus-like particle budding caused by HIV-1 protein Vpu. Eur Biophys J 31(1):26–35. https://doi.org/10.1007/s002490100177
Farha MA, Brown ED (2019) Drug repurposing for antimicrobial discovery. Nat Microbiol 4(4):565–577
Fehr AR, Perlman S (2015) Coronaviruses: an overview of their replication and pathogenesis. In Coronaviruses 1-23. In Perlman S, Gallagher T, Snijder E (ed), Nidoviruses. ASM Press, Washington, DC. https://doi.org/10.1128/9781555815790.ch12
Frediansyah A, Tiwari R, Sharun K, Dhama K, Harapan H (2020) Antivirals for COVID-19: a critical review. Clinical Epidemiology and global health 9:90–98. https://doi.org/10.1016/j.cegh.2020.07.006
Frieman M, Baric R (2008) Mechanisms of severe acute respiratory syndrome pathogenesis and innate immunomodulation. Microbiol Mol Biol Rev 72(4):672–685. https://doi.org/10.1128/mmbr.00015-08
Furuta Y, Komeno T, Nakamura T (2017) Favipiravir (T-705), a broad-spectrum inhibitor of viral RNA polymerase. Proc Jpn Acad Ser B 93(7):449–463. https://doi.org/10.2183/pjab.93.027
Gheblawi M, Wang K, Viveiros A, Nguyen Q, Zhong JC, Turner AJ, Raizada MK, Grant MB, Oudit GY (2020) Angiotensin-converting enzyme 2: SARS-CoV-2 receptor and regulator of the renin-angiotensin system: celebrating the 20th anniversary of the discovery of ACE2. Circ Res 126(10):1456–1474. https://doi.org/10.1161/circresaha.120.317015
Gioia M, Ciaccio C, De Simone G, Fasciglione GF, di Masi A, Di Pierro D, Bocedi A, Ascenzi P, Coletta M (2020) Role of proteolytic enzymes in the COVID-19 infection and promising therapeutic approaches. Biochem Pharmacol 182:114225. https://doi.org/10.1016/j.bcp.2020.114225
Griffin SD, Beales LP, Clarke DS, Worsfold O, Evans SD, Jaeger J, Harris MP, Rowlands DJ (2003) The p7 protein of hepatitis C virus forms an ion channel that is blocked by the antiviral drug. Amantadine FEBS letters 535(1–3):34–38. https://doi.org/10.1016/s0014-5793(02)03851-6
Guo YR, Cao QD, Hong ZS, Tan YY, Chen SD, ** HJ, Tan KS, Wang DY, Yan Y (2020) The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak–an update on the status. Mil Med Res 7(1):1. https://doi.org/10.1186/s40779-020-00240-0
Hamming I, Timens W, Bulthuis ML, Lely AT, Navis GJ, van Goor H (2004) Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. The Journal of Pathology: A Journal of the Pathological Society of Great Britain and Ireland 203(2):631–637. https://doi.org/10.1002/path.1570
Harrison C (2020) Coronavirus puts drug repurposing on the fast track. Nat Biotechnol 38(4):379–381. https://doi.org/10.1038/d41587-020-00003-1
Hilbert M, Kebbel F, Gubaev A, Klostermeier D (2011) eIF4G stimulates the activity of the DEAD box protein eIF4A by a conformational guidance mechanism. Nucleic Acids Res 39(6):2260–2270. https://doi.org/10.1093/nar/gkq1127
Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Müller MA (2020) SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181:271–280. https://doi.org/10.1016/j.cell.2020.02.052
Hofmann H, Pöhlmann S (2004) Cellular entry of the SARS coronavirus. Trends Microbiol 12(10):466–472. https://doi.org/10.1016/j.tim.2004.08.008
Hogue BG, Machamer CE (2007) Coronavirus structural proteins and virus assembly. Nidoviruses 179-200. https://doi.org/10.1128/9781555815790.ch12
Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z (2020) Clinical features of patients infected with 2019 novel coronavirus in Wuhan. China The lancet 395(10223):497–506. https://doi.org/10.1016/S0140-6736(20)30183-5
Huang Q, Yu L, Petros AM, Gunasekera A, Liu Z, Xu N, Hajduk P, Mack J, Fesik SW, Olejniczak ET (2004) Structure of the N-terminal RNA-binding domain of the SARS CoV nucleocapsid protein. Biochemistry 43(20):6059–6063. https://doi.org/10.1021/bi036155b
Hung AY, Sheng M (2002) PDZ domains: structural modules for protein complex assembly. J Biol Chem 277(8):5699–5702. https://doi.org/10.1074/jbc.r100065200
Jimenez-Guardeño JM, Nieto-Torres JL, DeDiego ML, Regla-Nava JA, Fernandez-Delgado R, Castaño-Rodriguez C, Enjuanes L (2014) The PDZ-binding motif of severe acute respiratory syndrome coronavirus envelope protein is a determinant of viral pathogenesis. PLoS Pathog 10(8):e1004320. https://doi.org/10.1371/journal.ppat.1004320
Kao RY, Tsui WH, Lee TS, Tanner JA, Watt RM, Huang JD, Hu L, Chen G, Chen Z, Zhang L, He T (2004) Identification of novel small-molecule inhibitors of severe acute respiratory syndrome-associated coronavirus by chemical genetics. Chem Biol 11(9):1293–1299. https://doi.org/10.1016/j.chembiol.2004.07.013
Karthik L, Kumar G, Keswani T, Bhattacharyya A, Chandar SS, Rao KB (2014) Protease inhibitors from marine actinobacteria as a potential source for antimalarial compound. PLoS ONE 9(3):90972. https://doi.org/10.1371/journal.pone.0090972
Kaur SP, Gupta V (2020) COVID-19 Vaccine: a comprehensive status report. Virus Res 288:198114. https://doi.org/10.1016/j.virusres.2020.198114
Kouznetsova VL, Zhang A, Tatineni M, Miller MA, Tsigelny IF (2020) Potential COVID-19 papain-like protease PLpro inhibitors: repurposing FDA-approved drugs. PeerJ 8:e9965. https://doi.org/10.7717/peerj.9965
Kuba K, Imai Y, Rao S, Gao H, Guo F, Guan B, Huan Y, Yang P, Zhang Y, Deng W, Bao L (2005) A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus–induced lung injury. Nat Med 11(8):875–879. https://doi.org/10.1038/nm1267
Kuo L, Hurst KR, Masters PS (2007) Exceptional flexibility in the sequence requirements for coronavirus small envelope protein function. J Virol 81(5):2249–2262. https://doi.org/10.1128/JVI.01577-06
Lamb YN, (2020) Remdesivir: first approval. Drugs 80(13):1355–1363. https://doi.org/10.1007/s40265-020-01378-w
Li F (2016) Structure, function, and evolution of coronavirus spike proteins. Annual review of virology 3:237–261. https://doi.org/10.1146/annurev-virology-110615-042301
Li H, Yang L, Liu FF, Ma XN, He PL, Tang W, Tong XK, Zuo JP (2020) Overview of therapeutic drug research for COVID-19 in China. Acta Pharmacologica Sinica 41(9):1133–40. https://www.x-mol.com/paperRedirect/1273356493890547712
Li Y, Surya W, Claudine S, Torres J (2014) Structure of a conserved Golgi complex-targeting signal in coronavirus envelope proteins. J Biol Chem 289(18):12535–12549. https://doi.org/10.1074/jbc.m114.560094
Lionta E, Spyrou G, Vassilatis K, D, Cournia Z, (2014) Structure-based virtual screening for drug discovery: principles, applications and recent advances. Curr Top Med Chem 14(16):1923–1938. https://doi.org/10.2174/1568026614666140929124445
Liu C, Zhu D (2020) Cyclophilin A and CD147: novel therapeutic targets for the treatment of COVID-19. Med Drug Discov 100056:1–8. https://doi.org/10.1016/j.medidd.2020.100056
Luo C, Luo H, Zheng S, Gui C, Yue L, Yu C, Sun T, He P, Chen J, Shen J, Luo X (2004) Nucleocapsid protein of SARS coronavirus tightly binds to human cyclophilin A. Biochem Biophys Res Commun 321(3):557–565. https://doi.org/10.1016/j.bbrc.2004.07.003
Madan V, de Jesús GM, Sanz MA, Carrasco L (2005) Viroporin activity of murine hepatitis virus E protein. FEBS Lett 579(17):3607–3612. https://doi.org/10.1016/j.febslet.2005.05.046
Matsuyama S, Nagata N, Shirato K, Kawase M, Takeda M, Taguchi F (2010) Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J Virol 84(24):12658–12664. https://doi.org/10.1128/jvi.01542-10
McBride R, Van Zyl M, Fielding BC (2014) The coronavirus nucleocapsid is a multifunctional protein. Viruses 6(8):2991–3018. https://doi.org/10.3390/v6082991
Millet JK, Whittaker GR (2014) Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc Natl Acad Sci 111(42):15214–15219. https://doi.org/10.1073/pnas.1407087111
Mittal A, Manjunath K, Ranjan RK, Kaushik S, Kumar S, Verma V (2020) COVID-19 pandemic: Insights into structure, function, and hACE2 receptor recognition by SARS-CoV-2. PLoS Pathog 16(8):e1008762. https://doi.org/10.1371/journal.ppat.1008762
Momattin H, Mohammed K, Zumla A, Memish ZA, Al-Tawfiq JA (2013) Therapeutic options for Middle East respiratory syndrome coronavirus (MERS-CoV)–possible lessons from a systematic review of SARS-CoV therapy. Int J Infect Dis 17(10):e792–e798
Montero H, Pérez-Gil G, Sampieri CL (2019) Eukaryotic initiation factor 4A (eIF4A) during viral infections. Virus Genes 55(3):267–273. https://doi.org/10.1007/s11262-019-01641-7
Mousavizadeh L, Ghasemi S (2020) Genotype and phenotype of COVID-19: their roles in pathogenesis. J Microbiol Immunol Infect 54:159–163. https://doi.org/10.1016/j.jmii.2020.03.022
Müller C, Schulte FW, Lange-Grünweller K, Obermann W, Madhugiri R, Pleschka S, Ziebuhr J, Hartmann RK, Grünweller A (2018) Broad-spectrum antiviral activity of the eIF4A inhibitor silvestrol against corona-and picornaviruses. Antiviral Res 150:123–129. https://doi.org/10.1016/j.antiviral.2017.12.010
Muralidharan N, Sakthivel R, Velmurugan D, Gromiha MM (2020) Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 protease against COVID-19. J Biomol Struct Dynamic 39(7):2673–2678. https://doi.org/10.1080/07391102.2020.1752802
Nakagawa K, Lokugamage KG, Makino S (2016) Viral and cellular mRNA translation in coronavirus-infected cells. Adv Virus Res 96:165–192. https://doi.org/10.1016/bs.aivir.2016.08.001
Nebigil CG, Moog C, Vagner S, Benkirane-Jessel N, Smith DR, Désaubry L (2020) Flavaglines as natural products targeting eIF4A and prohibitins: from traditional Chinese medicine to antiviral activity against coronaviruses. Eur J Med Chem 203:112653. https://doi.org/10.1016/j.ejmech.2020.112653
Neuman BW (2016) Bioinformatics and functional analyses of coronavirus nonstructural proteins involved in the formation of replicative organelles. Antiviral Res 135:97–107. https://doi.org/10.1016/j.antiviral.2016.10.005
O’Keefe B, Giomarelli B, Barnard DL, Shenoy SR, Chan P, McMahon JB, Palmer KE, Barnett BW, Meyerholz DK, Wohlford-Lenane CL, McCray PB Jr (2010) Broad-spectrum in vitro activity and in vivo efficacy of the antiviral protein griffithsin against emerging viruses of the family Coronaviridae. J Virol 84(5):2511–2521. https://doi.org/10.1128/jvi.02322-09
O’Meara MJ, Guo JZ, Swaney DL, Tummino TA, Hüttenhain R (2020) A SARS-CoV-2-human protein-protein interaction map reveals drug targets and potential drug-repurposing. BioRxiv 1–45. https://doi.org/10.1038/s41586-020-2286-9
Ortega JT, Serrano ML, Pujol FH, Rangel HR (2020) Role of changes in SARS-CoV-2 spike protein in the interaction with the human ACE2 receptor: an in-silico analysis. EXCLI J 19:410–417. https://doi.org/10.17179/excli2020-1167
Othman H, Bouslama Z, Brandenburg JT, Da Rocha J, Hamdi Y, Ghedira K, Srairi-Abid N, Hazelhurst S (2020) Interaction of the spike protein RBD from SARS-CoV-2 with ACE2: similarity with SARS-CoV, hot-spot analysis and effect of the receptor polymorphism. Biochem Biophys Res Commun 527(3):702–708. https://doi.org/10.1016/j.bbrc.2020.05.028
Peng Q, Peng R, Yuan B, Zhao J, Wang M, Wang X, Wang Q, Sun Y, Fan Z, Qi J, Gao GF (2020) Structural and biochemical characterization of the nsp12-nsp7-nsp8 core polymerase complex from SARS-CoV-2. Cell Rep 31(11):107774
Perrotta F, Matera MG, Cazzola M, Bianco A (2020) Severe respiratory SARS-CoV2 infection: does ACE2 receptor matter? Respir Med 168:105996. https://doi.org/10.1016/j.rmed.2020.105996
Prabakaran P, Gan J, Feng Y, Zhu Z, Choudhry V, **ao X, Ji X, Dimitrov DS (2006) Structure of severe acute respiratory syndrome coronavirus receptor-binding domain complexed with neutralizing antibody. J Biol Chem 281(23):15829–15836. https://doi.org/10.1074/jbc.m600697200
Prabhu SA, Moussa O, Miller WH, del Rincón SV (2020) The MNK1/2-eIF4E axis as a potential therapeutic target in melanoma. Int J Mol Sci 21(11):4055. https://doi.org/10.3390/ijms21114055
Prajapat M, Sarma P, Shekhar N, Avti P, Sinha S, Kaur H, Kumar S, Bhattacharyya A, Kumar H, Bansal S, Medhi B (2020) Drug targets for corona virus: a systematic review. Indian J Pharmacol 52(1):56. https://doi.org/10.4103/ijp.IJP_115_20
Quimque MT, Notarte KI, Fernandez RA, Mendoza MA, Liman RA, Lim JA, Pilapil LA, Ong JK, Pastrana AM, Khan A, Wei DQ (2020). Virtual screening-driven drug discovery of SARS-CoV2 enzyme inhibitors targeting viral attachment, replication, post-translational modification and host immunity evasion infection mechanisms. J Biomol Struct Dynam 1-18. https://doi.org/10.1080/07391102.2020.1776639
Raj K, Rohit AG, Singh S (2020) Coronavirus as silent killer: recent advancement to pathogenesis, therapeutic strategy and future perspectives. VirusDisease 2020:1–9. https://doi.org/10.1007/s13337-020-00580-4
Rajiv C, Davis TL (2018) Structural and functional insights into human nuclear cyclophilins. Biomolecules 8(4):161. https://doi.org/10.3390/biom8040161
Raoult D, Zumla A, Locatelli F, Ippolito G, Kroemer G (2020) Coronavirus infections: epidemiological, clinical and immunological features and hypotheses. Cell Stress 4(4):66. https://doi.org/10.15698/cst2020.04.216
Robson B (2020) COVID-19 Coronavirus spike protein analysis for synthetic vaccines, a peptidomimetic antagonist, and therapeutic drugs, and analysis of a proposed achilles’ heel conserved region to minimize probability of escape mutations and drug resistance. Comput Biol Med 103749:1–28. https://doi.org/10.1016/j.compbiomed.2020.103749
Sangawa H, Komeno T, Nishikawa H, Yoshida A, Takahashi K, Nomura N, Furuta Y (2013) Mechanism of action of T-705 ribosyl triphosphate against influenza virus RNA polymerase. Antimicrob Agents Chemother 57(11):5202–5208. https://doi.org/10.1128/aac.00649-13
Senanayake SL (2020) Drug repurposing strategies for COVID-19 2(2):1–3. https://doi.org/10.4155/fdd-2020-0010
Shah VK, Firmal P, Alam A, Ganguly D, Chattopadhyay S (2020) Overview of immune response during SARS-CoV-2 infection: lessons from the past. Front Immunol 11:1949. https://doi.org/10.3389/fimmu.2020.01949
Shang J, Wan Y, Luo C, Ye G, Geng Q, Auerbach A, Li F (2020) Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci 117(21):11727–11734. https://doi.org/10.1073/pnas.2003138117
Shulla A, Heald-Sargent T, Subramanya G, Zhao J, Perlman S, Gallagher T (2011) A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J Virol 85(2):873–882. https://doi.org/10.1128/jvi.02062-10
Simmons G, Zmora P, Gierer S, Heurich A, Pöhlmann S (2013) Proteolytic activation of the SARS-coronavirus spike protein: cutting enzymes at the cutting edge of antiviral research. Antiviral Res 100(3):605–614. https://doi.org/10.1016/j.antiviral.2013.09.028
Singh TU, Parida S, Lingaraju MC, Kesavan M, Kumar D, Singh RK (2020) Drug repurposing approach to fight COVID-19. Pharmacological Reports 72:1479–1508. https://doi.org/10.1007/s43440-020-00155-6
Skehel JJ, Hay AJ, Armstrong JA (1978) On the mechanism of inhibition of influenza virus replication by amantadine hydrochloride. J Gen Virol 38(1):97–110. https://doi.org/10.1099/0022-1317-38-1-97
Slaine PD, Kleer M, Smith NK, Khaperskyy DA, McCormick C (2017) Stress granule-inducing eukaryotic translation initiation factor 4A inhibitors block influenza A virus replication. Viruses 9(12):388. https://doi.org/10.3390/v9120388
Sola I, Almazan F, Zuniga S, Enjuanes L (2015) Continuous and discontinuous RNA synthesis in coronaviruses. Annu Rev Virol 2:265–288. https://doi.org/10.1146/annurev-virology-100114-055218
Song Z, Yang Y, Wang L, Wang K, Ran L, **e Y, Huang L, Yang Z, Yuan P, Yu Q (2019) EIF4A2 interacts with the membrane protein of transmissible gastroenteritis coronavirus and plays a role in virus replication. Res Vet Sci 123:39–46. https://doi.org/10.1016/j.rvsc.2018.12.005
Sparks MA, Crowley SD, Gurley SB, Mirotsou M, Coffman TM (2011) Classical renin-angiotensin system in kidney physiology. Compr Physiol 4(3):1201–1228. https://doi.org/10.1002/cphy.c130040
Stockman LJ, Bellamy R, Garner P (2006) SARS: systematic review of treatment effects. PLoS med 3(9):e343. https://doi.org/10.1371/journal.pmed.0030343
Surjit M, Lal SK (2008) The SARS-CoV nucleocapsid protein: a protein with multifarious activities. Infect Genet Evol 8(4):397–405. https://doi.org/10.1016/j.meegid.2007.07.004
Talluri S (2020) Molecular docking and virtual screening based prediction of drugs for COVID-19. Comb Chem High Throughput Screen. https://doi.org/10.2174/1386207323666200814132149
Tanaka Y, Sato Y, Sasaki T (2017) Feline coronavirus replication is affected by both cyclophilin A and cyclophilin B. J Gen Virol 98(2):190–200. https://doi.org/10.1099/jgv.0.000663
Taroncher-Oldenburg G, Müller C, Obermann W, Ziebuhr J, Hartmann RK, Grünweller (2021) A. Targeting the DEAD-box RNA helicase eIF4A with Rocaglates-A Pan-antiviral strategy for minimizing the impact of future RNA virus Pandemics. Microorganisms 9(3):540–558. https://doi.org/10.20944/preprints202102.0058.v1
Teoh KT, Siu YL, Chan WL, Schlüter MA, Liu CJ, Peiris JM, Bruzzone R, Margolis B, Nal B (2010) The SARS coronavirus E protein interacts with PALS1 and alters tight junction formation and epithelial morphogenesis. Mol Biol Cell 21(22):3838–3852. https://doi.org/10.1091/mbc.e10-04-0338
Thompson PA, Eam B, Young NP, Fish S, Chen J, Barrera M, Howard H, Sung E, Parra A, Staunton J, Chiang GG (2019) eFT226, a potent and selective inhibitor of eIF4A, is efficacious in preclinical models of lymphoma. 79(13):2698–2698. https://doi.org/10.1099/jgv.0.000663
Tian L, Qiang T, Liang C, Ren X, Jia M, Zhang J, Li J, Wan M, YuWen X, Li H, Cao W (2021a) RNA-dependent RNA polymerase (RdRp) inhibitors: the current landscape and repurposing for the COVID-19 pandemic. Eur J Med Chem 213:113201. https://doi.org/10.1016/j.ejmech.2021.113201
Torres J, Maheswari U, Parthasarathy K, Ng L, Liu DX, Gong X (2007) Conductance and amantadine binding of a pore formed by a lysine-flanked transmembrane domain of SARS coronavirus envelope protein. Protein Sci 16(9):2065–2071. https://doi.org/10.1110/ps.062730007
Touret F, Gilles M, Barral K, Nougairède A, van Helden J, Decroly E, de Lamballerie X, Coutard B (2020) In vitro screening of a FDA approved chemical library reveals potential inhibitors of SARS-CoV-2 replication. Sci Rep 10(1):1–8. https://doi.org/10.1038/s41598-020-70143-6
Tsumuraya T, Ishikawa C, Machijima Y, Nakachi S, Senba M, Tanaka J, Mori N (2011) Effects of hippuristanol, an inhibitor of eIF4A, on adult T-cell leukemia. Biochem Pharmacol 81(6):713–722. https://doi.org/10.1016/j.bcp.2010.12.025
ul Qamar MT, Alqahtani SM, Alamri MA, Chen LL (2020) Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants. J Pharmaceut Analy. 2020:1-7 https://doi.org/10.1016/j.jpha.2020.03.009
Ulferts R, Imbert I, Canard B, Ziebuhr J (2010) Expression and functions of SARS coronavirus replicative proteins. In Molecular biology of the SARS-coronavirus. Springer, Berlin, Heidelberg 75–98. https://doi.org/10.1007/978-3-642-03683-5_6
van Boheemen S, de Graaf M, Lauber C, Bestebroer TM, Raj VS, Zaki AM, Osterhaus AD, Haagmans BL, Gorbalenya AE, Snijder EJ, Fouchier RA (2012) Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. MBio 3(6):e00473-12. https://doi.org/10.1128/mbio.00473-12
von Hahn T, Ciesek S (2015) Cyclophilin polymorphism and virus infection. Curr Opin Virol 14:47–49. https://doi.org/10.1016/j.coviro.2015.07.012
Wang Y, Anirudhan V, Du R, Cui Q, Rong L (2021) RNA-dependent RNA polymerase of SARS-CoV-2 as a therapeutic target. J Med Virol 93(1):300–310. https://doi.org/10.1002/jmv.26264
Wang Y, Li W, Jiang Z, ** X, Zhu Y (2020) Assessment of the efficacy and safety of Ribavirin in treatment of coronavirus-related pneumonia (SARS, MERS and COVID-19): a protocol for systematic review and meta-analysis. Medicine 99(38):e22379. https://doi.org/10.1097/md.0000000000022379
Wang YS, Chen J, Cui F, Wang H, Wang S, Hang W, Zeng Q, Quan CS, Zhai YX, Wang JW, Shen XF (2016) LKB1 is a DNA damage response protein that regulates cellular sensitivity to PARP inhibitors. Oncotarget 7(45):73389. https://doi.org/10.18632/oncotarget.12334
White KM, Rosales R, Yildiz S, Kehrer T, Miorin L, Moreno E, Jangra S, Uccellini MB, Rathnasinghe R, Coughlan L, Martinez-Romero C (2021) Plitidepsin has potent preclinical efficacy against SARS-CoV-2 by targeting the host protein eEF1A. Science 371(6532):926–931. https://doi.org/10.1126/science.abf4058
Wilson L, Gage P, Ewart G (2006) Hexamethylene amiloride blocks E protein ion channels and inhibits coronavirus replication. Virology 353(2):294–306. https://doi.org/10.1016/j.virol.2006.05.028
Wilson L, Mckinlay C, Gage P, Ewart G (2004) SARS coronavirus E protein forms cation-selective ion channels. Virology 330(1):322–331. https://doi.org/10.1016/j.virol.2004.09.033
Wondmkun YT, Mohammed OA (2020) A review on novel drug targets and future directions for COVID-19 treatment. Biologics: Targets & Therapy 14(77):77–82
Wu CJ, Huang HW, Liu CY, Hong CF, Chan YL (2005) Inhibition of SARS-CoV replication by siRNA. Antiviral Res 65(1):45–8. https://doi.org/10.1016/j.antiviral.2004.09.005
Wu R, Wang L, Kuo HC, Shannar A, Peter R, Chou PJ, Li S, Hudlikar R, Liu X, Liu Z, Poiani GJ (2020) An update on current therapeutic drugs treating COVID-19. Curr Pharmacol Rep 6(3): 56–70. https://doi.org/10.1007/s40495-020-00216-7
Ye Y, Hogue BG (2007) Role of the coronavirus E viroporin protein transmembrane domain in virus assembly. J Virol 81(7):3597–3607. https://doi.org/10.1128/jvi.01472-06
Zhang J, **e B, Hashimoto K (2020) Current status of potential therapeutic candidates for the COVID-19 crisis. Brain, Behavior, and Immunity 1–15. https://doi.org/10.1016/j.bbi.2020.04.046
Zhang T, Wu Q, Zhang Z (2020) Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Curr Biol 30(7):1346–51. https://doi.org/10.1016/j.cub.2020.03.022
Zhou D, Mei Q, Li J, He H (2012) Cyclophilin A and viral infections. Biochem Biophys Res Commun 424(4):647–650. https://doi.org/10.1016/j.bbrc.2012.07.024
Zhou Y, Hou Y, Shen J, Huang Y, Martin W, Cheng F (2020) Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell discovery 6(1):1–8. https://doi.org/10.1038/s41421-020-0153-3
Ziebuhr J (2005) The coronavirus replicase. Coronavirus replication and reverse genetics. Springer, Berlin, pp 57–94
Author information
Authors and Affiliations
Contributions
Dr. SS and Prof. GD designed, drafted, edited, and corrected grammatical errors in the revised manuscript. KR and KK carried out the literature review and written the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethical approval
This article is a review article, so it does not contain any studies with human participants performed by any of the authors.
Consent to participate
Not applicable.
Consent to publish
Not applicable.
Competing of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Raj, K., Kaur, K., Gupta, G.D. et al. Current understanding on molecular drug targets and emerging treatment strategy for novel coronavirus-19. Naunyn-Schmiedeberg's Arch Pharmacol 394, 1383–1402 (2021). https://doi.org/10.1007/s00210-021-02091-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00210-021-02091-5