Abstract
Metformin is the first-line drug treatment for type 2 diabetes. Globally, over 100 million patients are prescribed this drug annually. Metformin was discovered before the era of target-based drug discovery and its molecular mechanism of action remains an area of vigorous diabetes research. An improvement in our understanding of metformin’s molecular targets is likely to enable target-based identification of second-generation drugs with similar properties, a development that has been impossible up to now. The notion that 5' AMP-activated protein kinase (AMPK) mediates the anti-hyperglycaemic action of metformin has recently been challenged by genetic loss-of-function studies, thrusting the AMPK-independent effects of the drug into the spotlight for the first time in more than a decade. Key AMPK-independent effects of the drug include the mitochondrial actions that have been known for many years and which are still thought to be the primary site of action of metformin. Coupled with recent evidence of AMPK-independent effects on the counter-regulatory hormone glucagon, new paradigms of AMPK-independent drug action are beginning to take shape. In this review we summarise the recent research developments on the molecular action of metformin.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
According to national and international guidelines, metformin is the recommended first-line oral therapy for the treatment of type 2 diabetes [1–3]. This is down to several factors, including the impressive safety record of the drug, having been in clinical use for over 50 years and the fact that metformin treatment is weight neutral. In addition, there are likely to be other beneficial effects, including a reduction in cardiovascular disease and mortality compared with non-intensive treatment [4] and a possible reduction in cancer incidence, which has been seen in some [5–7], but not all [8], studies. As metformin was discovered in the era before modern target-based drug discovery, the molecular details of its mechanism of action were not established before it was used clinically and these continue to be an area of vigorous research. In this review we discuss what is known and unknown about the molecular action of metformin.
Molecular action and target of metformin: lessons and insights from early studies
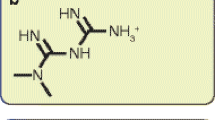
Chemically, biguanides such as metformin are composed of two guanidine groups joined together with the loss of ammonia. Anti-hyperglycaemic effects have been observed in response to many, but not all, guanidine-containing compounds. For metformin, these effects are uniquely dissociated from toxicity. Before the biguanides, attention was first focused on guanidine itself [9], which was too toxic for clinical use, then diguanides (also known as synthalins or diguanidines) [10], composed of two guanidines connected by an alkyl chain of variable length. Two diguanides, synthalin A and synthalin B, were used clinically, but marked toxicity, which could not be dissociated from therapeutic effects, was noted quite quickly [11, 12]. By the late 1950s, attention shifted to metformin and two other biguanides, phenformin and buformin. Even amongst these biguanides, metformin exhibits a superior safety profile. This is primarily because the risk of lactic acidosis, which can be fatal, is much higher for phenformin or buformin treatment [13].
The large quantities of the drug required (up to 2.5 g per day) for therapeutic effects led early investigators to hypothesise that it might not depend on a conventional single/specific protein target. Early physiological studies on the diguanides found that reduced oxygen consumption accompanied hypoglycaemia [14]. Later work found that guanidine [15, 16], diguanides [16] and phenformin [17, 18] reduce mitochondrial oxygen consumption, suggesting that this organelle is an important site of action of guanidine-based agents. Several models have been proposed to account for these effects. The first model was based on observations that a common property of biguanides and other guanidines is to change the charge distribution and/or net charge of membranes [19]. This work showed that these drugs could inhibit the transport of protons and other cations across membranes [19]. However, the magnitude of these effects did not correlate well with anti-hyperglycaemic efficacy, with some ineffective drugs interacting with membranes much more readily than either phenformin or metformin [20]. Furthermore, whilst phenformin has appreciable hydrophobicity and interacts with membranes, metformin is unusually hydrophilic for a drug and is unlikely to interact with membranes significantly. For this reason, metformin is understood to require transporters to cross membranes. Another suggestion during this period was that guanidine-containing drugs induce anti-hyperglycaemic effects by displacing calcium from proteins such as pyruvate kinase [21]; however, as in the experiments on membranes, these effects occurred only at very high concentrations and hydrophobicity was an important determinant of potency. Thus, the IC50 by phenformin was 2.5 mmol/l, but the corresponding value for metformin was 275 mmol/l—far higher even than the non-specific cation tetramethylammonium [22]. This suggests that the calcium-mobilising effects described are unlikely to contribute to the therapeutic action of metformin either. Whether or not calcium-mobilising or lipid effects contribute to the toxicity of guanidines such as phenformin has not been thoroughly investigated. It has recently been suggested that the effects of metformin on the mitochondria depend on a third non-protein effect—direct targeting of metal ions [23]—which takes the form of an unusual electron delocalised planar ring structure, where square planar geometry replaces more conventional tetragonal geometry [24]. The compelling evidence of direct binding of metformin to metal ions, including extensive crystallographic [25] and spectroscopic analysis [24, 26–28], contrasts with the paucity of evidence regarding direct binding of the drug to recognised metformin-regulated proteins. Further work is required to establish how the metal-binding properties of metformin enable it to mediate mitochondrial inhibition.
A second important physiological response to biguanides is reduced gluconeogenesis [29]. Over time, studies on guanidine derivatives began to link inhibition of mitochondrial respiration with reduced gluconeogenesis [30]; however, as already indicated, the poor correlation of the magnitudes of these effects for some drugs led others to conclude that mitochondrial effects were more likely to contribute towards side effects such as lactic acidosis rather than therapeutic effects [19, 21]. In the case of metformin, for example, there is often little impact on cellular ATP levels, even when using concentrations well above those likely to be achieved in vivo. These difficulties led to the suggestion that anti-hyperglycaemic effects might owe more to drug-specific mitochondrial effects superimposed upon the general mitochondrial responses to guanide-containing drugs already described [19]. One such effect of metformin was found in 2000, in a study of its effects on electron transport, the mitochondrial oxygen-dependent process that couples the citric acid cycle to ATP production, providing the bulk of most cells’ energy requirements (Fig. 1). Studies employing hepatocytes, mitochondria and freeze-clamped livers found that metformin’s suppression of hepatic glucose output is accompanied by inhibition of complex I in the mitochondrial electron transport chain [31, 32]. One key piece of evidence implicating complex I as the site of metformin action was that the drug inhibited mitochondrial oxidation of glutamate and malate more effectively than succinate [31, 32], which as a complex II substrate, can bypass complex I inhibition. Similar observations had been made earlier using other guanide-containing drugs [15, 16]; however, our understanding of mitochondrial respiration was probably insufficiently developed to allow this interpretation of the results at the time. Taken together, these studies provide compelling evidence of a correlation between inhibition of electron transport and glucose output. It should be noted, however, that it has not yet been possible to confirm in genetic experiments whether or not complex I is the only mitochondrial target of metformin. For example, it may be significant that recent studies have found that effects of metformin on mitochondrial respiration vary between cells, but more work is required to understand the underlying reason(s) for these variations [33].
(a, b) Schematic diagram of the anti-hyperglycaemic action of metformin on the liver cell. Part (b) shows a simplified version of (a). Metformin is transported into hepatocytes mainly via OCT1, resulting in an inhibition of the mitochondrial respiratory chain (complex I) through a currently unknown mechanism(s). The resulting deficit in energy production is balanced by reducing the consumption of energy in the cell, particularly reduced gluconeogenesis in the liver. This is mediated in two main ways. First, a decrease in ATP and a concomitant increase in AMP concentration occur, which is thought to contribute to the inhibition of gluconeogenesis directly (because of the energy/ATP deficit). Second, increased AMP levels function as a key signalling mediator that has been proposed to (1) allosterically inhibit cAMP–PKA signalling through suppression of adenylate cyclase, (2) allosterically inhibit FBPase, a key gluconeogenic enzyme, and (3) activates AMPK. This leads to inhibition of gluconeogenesis (1 and 2) and lipid/cholesterol synthesis (3), which may contribute to the longer term metabolic and therapeutic responses to the drug. FBPase; fructose-1,6-bisphosphatase
Studies on mitochondrial responses to metformin have reported that the magnitude of inhibition of gluconeogenesis is correlated to the extent of inhibition of the respiratory chain [31]. This suggests that metformin-dependent cellular energy depletion results in insufficient flux of ATP required to drive energy-consuming hepatic gluconeogenesis (Fig. 1) [31]. ATP-/ADP-/AMP-independent effects on pyruvate carboxylase and other enzymes may also contribute to the inhibition of gluconeogenesis accompanying mild mitochondrial inhibition [31, 34]. Mitochondrial inhibition also provides a plausible explanation of lactic acidosis in metformin use, as lactate accumulation from glycolysis would be predicted to follow respiratory chain inhibition. Compared with phenformin and buformin, lactic acidosis is very much less common with metformin therapy [35–37]. This might be because metformin-dependent mitochondrial inhibition is understood to be self-limiting as it depends on the mitochondria being active [31]. This property might also explain why metformin is very much less toxic than other complex I inhibitors, including the neurotoxic pesticide rotenone [31]. Interestingly, thiazolidinediones, another class of drugs used for the treatment of type 2 diabetes, are also mild inhibitors of complex I, and this might contribute to their anti-hyperglycaemic effect [38].
LKB1–AMPK-dependent pathway as a key molecular effector of metformin
A key study by Zhou et al in 2001 reported a ‘modern’ signal transduction effect of metformin on 5' AMP-activated protein kinase (AMPK) [39] (Fig. 1). AMPK is a critical cellular energy sensor and regulator of energy homeostasis [40]. AMPK is activated by energy stresses that increase cellular ADP:ATP and/or AMP:ATP ratios, either by decreasing the catabolic production of ATP (e.g. via nutrient deprivation and exposure to mitochondrial toxins) or by promoting ATP consumption (e.g. by muscle contraction). Zhou et al [39] hypothesised that AMPK might be an important molecular effector, as (1) metformin causes a reduction in cellular ATP:ADP ratios in hepatocytes [31, 32]; and (2) downstream effects of AMPK activation (e.g. promotion of glucose uptake and fatty acid oxidation in muscle, inhibition of lipid synthesis in the liver [41]) simulate the therapeutic effects of metformin. They demonstrated that metformin indeed stimulates AMPK, and this stimulation is associated with inhibition of glucose production in rat primary hepatocytes. They also showed that metformin treatment decreases levels of sterol regulatory element-binding protein-1 (SREBP-1), a key lipogenic transcription factor, at both the mRNA and protein level in hepatocytes and liver tissue [39]. In addition, they reported that metformin increases fatty acid oxidation in hepatocytes, although oral ingestion of metformin resulted in suppression of whole body lipid oxidation as well as inhibition of hepatic glucose production in humans [42]. To demonstrate that the observed effects of metformin are sensitive to changes in the AMPK-dependent pathway, Zhou et al used a novel small molecule AMPK inhibitor named compound C. When rat primary hepatocytes were incubated with compound C, metformin-induced inhibition of acetyl-CoA carboxylase (a well known target of AMPK involved in lipid metabolism) and inhibition of glucagon-induced glucose production were significantly attenuated [39]. However, it should be noted that several recent publications have shown that, in addition to AMPK, compound C potently and non-selectively inhibits numerous other protein kinases and therefore the results obtained using compound C are not conclusive [43, 44]. It has been shown that metformin does not directly target AMPK or affect its phosphorylation by upstream kinases (and phosphatases) in cell-free systems. A recent study has provided compelling evidence that it activates AMPK in an indirect manner via an increase in AMP:ATP and ADP:ATP ratios using an engineered cell line expressing AMPK complexes bearing either the wild-type γ2 isoform or Arg531 → Gly mutation that renders γ2 complexes insensitive to the effects of ADP and AMP on phosphorylation [33].
The proposed key role for AMPK in mediating metformin action was further followed up by Shaw et al in 2005. They sought to determine the role of tumour suppressor protein LKB1, an upstream kinase of AMPK, in the liver and generated a liver-specific Lkb1 (also known as Stk11) knockout mouse model [45]. They found that metformin-induced AMPK activation was profoundly reduced in Lkb1 knockout liver and that, strikingly, metformin treatment failed to produce a glucose-lowering effect in liver-specific Lkb1 knockout mice rendered hyperglycaemic (by feeding these mice a fat-enriched diet). Shaw et al proposed that the LKB1–AMPK signalling controls the expression of key gluconeogenic genes via the regulation of a transcription coactivator [45], namely, cAMP response element-binding protein-regulated transcription coactivator 2 (CRTC2) [46]. In the fasting state, CRTC2 is in a dephosphorylated state and is localised in the nucleus where it enhances the transcriptional activation of the gluconeogenic genes, including those encoding peroxisome proliferator-activated receptor-γ coactivator-1α (Ppargc1a) and its subsequent gluconeogenic targets such as phosphoenolpyruvate carboxykinase (Pck1) and glucose-6 phosphatase (G6pc). Feeding (insulin) is thought to switch off glucose production (at least partially) through inhibition of gluconeogenic gene programmes via Akt kinase-signalling-dependent phosphorylation and cytoplasmic sequestration of CRTC2 and forkhead box O1 (FOXO1) proteins [47]. Interestingly, metformin can also switch off gluconeogenesis independently of insulin/Akt signalling via LKB1–AMPK pathway. Consistent with this model, in fasted liver-specific Lkb1 knockout mice, CRTC2 was localised predominantly in the nucleus, which was associated with increases in Ppargc1a and G6pc gene expression, as well as blood glucose levels compared with control mice [45].
To establish whether the effects of metformin on glucoeneogenesis in the liver are mainly mediated through AMPK or LKB1-dependent kinases (other than AMPK), Foretz et al [48] recently generated and examined two mouse models in which genes encoding the AMPKα1 and α2 catalytic subunits (Prkaa1/2) or LKB1 are knocked out specifically in hepatocytes. Interestingly, they showed that the administration of a bolus injection of metformin to hepatocyte-specific Prkaa1/2 knockout mice resulted in a glucose-lowering effect comparable with that observed in control mice. In addition, treatment of primary hepatocytes lacking AMPK with metformin displayed a robust inhibition of glucose production (induced via the membrane-permeable cAMP analogue dibutyryl cAMP, which mimics the action of glucagon) and, in fact, the magnitude of inhibition was even greater than that observed in control wild-type hepatocytes [48]. Consistent with the observation that liver-specific Lkb1 knockout mice display higher blood glucose levels in the fasting state [45], Foretz et al reported that Lkb1 −/− hepatocytes have higher rates of basal/unstimulated and cAMP-stimulated glucose production compared with Lkb1 +/+ cells. However, unexpectedly, Foretz et al demonstrated that metformin inhibits glucose production normally in Lkb1 knockout hepatocytes [48]. One potential explanation for this discrepancy is that Shaw et al did not assess the immediate effect of metformin administration on hepatic glucose output by their liver-specific Lkb1 knockout mice, either in vivo or in vitro (primary hepatocytes) [45]. Instead, they reported that the protective effect of metformin against hyperglycaemia in response to the fat-enriched diet was eliminated by repeated administration (daily injection) of metformin in liver-specific Lkb1 knockout mice. Thus, it can be speculated that, in the study of Shaw et al, metformin did not act directly to suppress glucose production but, rather, might have acted indirectly to protect the hepatocytes from high-fat diet-induced lipotoxicity and associated insulin resistance through suppression of lipid synthesis/lipogenic gene expression (Fig. 1). Finally, it should be noted that, although the mouse genetic model is a useful tool, inactivation of critical metabolic gene(s) could possibly result in compensatory adaptations in alternative pathway(s) to maintain glucose homeostasis and, in addition, results from mouse studies are not always applicable to human physiology and pathology. To date, there is no literature reporting that patients who have a poor glycaemic response to metformin have impaired AMPK signalling. There are many loss-of-function mutations of LKB1 associated with Peutz–Jeghers cancer syndrome [49]; however, to the best of our knowledge, these patients have not been found to have a higher incidence of develo** metabolic syndromes such as type 2 diabetes or insulin resistance, and Lkb1 variants have not convincingly been reported to play a role in the glycaemic response to metformin in type 2 diabetes.
The liver is the main site of action of metformin: the role of organic cation transporters
Metformin is not metabolised and is excreted in the urine and bile in an unmodified form. The pharmacokinetics of metformin are largely determined by its active transport by key organic cation transporters. Members of this transporter family [50] are involved in active transport across the gut epithelium and hence determine rates of absorption (plasma membrane monoamine transporter [PMAT] and organic cation transporter [OCT] 3), they transport metformin into hepatocytes (OCT1) and from hepatocytes into the bile (multidrug and toxic compound extrusion [MATE] 1) and, finally, into the renal tubular epithelial cells (OCT2) and into the renal tubule (MATE2) [51]. Oct1 (also known as Slc22a1) knockout mice display reduced efficacy of metformin [52] and this work has established an important role for OCT1 in metformin intake and reinforces the critical role of the liver as the primary site of action for metformin.
Although metformin exerts its major effect through inhibition of hepatic glucose production, enhanced glucose disposal has also been described. Some early studies suggested that metformin exerts its insulin-sensitising effect [53] and/or promotes glucose transport independently of the insulin receptor-mediated proximal signalling pathway in skeletal muscle [39, 54, 55]. Even though metformin may accumulate in skeletal muscle and other organs/tissues over longer periods of time to bring about some effects [56], the concentration of metformin that was required to acutely stimulate AMPK and glucose transport in isolated rodent muscle tissue ex vivo [39] or in cultured muscle cells [54] was at least two orders of magnitude greater (typically used at ∼1–2 mmol/l and treated for 3–16 h) than those seen in plasma following the administration of therapeutic doses (mean plasma concentration of 4.5 μmol/l) [57, 58], making it unlikely that metformin has a major therapeutic effect in the muscle. The potent and preferential effects of metformin in the liver can be explained by the fact that the drug is supplied directly from the gut (via the portal vein), which means that a profoundly higher concentration of metformin (when taken orally) reaches the liver than other peripheral organs/tissues [59], and by the high level of expression of OCT1, which actively transports metformin to its site of action, in hepatocytes. Metformin has also been reported to have a protective effect on the vascular endothelium, possibly explaining the potential cardiovascular benefit of this drug. This might reflect an indirect benefit owing to a reduction in hepatic glucose output and circulating insulin, but might also reflect a direct action of metformin on vascular endothelial cells, possibly activating AMPK and thereby increasing nitric oxide synthesis [60, 84]. In addition, type 2 diabetes is a progressive disorder that necessitates additional medication following failure of metformin monotherapy [1]. Progress in establishing the identity of the clinically important target(s) and key effectors of metformin will ultimately enable rational target-based design of more potent or better-tolerated drugs to support metformin use. For example, if post-glucagon receptor modification of glucagon signalling, as reported by Miller et al for phenformin treatment in rodents [67], can be shown to be an important effector of metformin therapy in humans, the allosteric inhibitor P-site on adenylate cyclase provides an alternative approach to blocking the receptor itself, and this offers an intriguing novel mechanism for inhibiting glucagon action in the liver. There are glucagon receptor antagonists in development and these offer promise as potential therapeutic agents [73]. However, there is some concern that direct blockade of the receptor may result in islet hyperplasia, as seen in rodents in which the glucagon receptor has been knocked out [72]. Additionally, increases in serum cholesterol and transaminases have been reported for glucagon receptor antagonists in clinical trial; effects not seen with metformin [85]. Whether this reflects its post-glucagon receptor action or other mechanisms of metformin is unclear. An alternative approach based on work on the metal-binding properties of metformin would be to develop agents capable of directly targeting copper ion transport or exchange. Type 2 diabetes is known to be accompanied by altered copper handling [86, 87], and work with other drugs has already demonstrated that drug-based copper sequestration ameliorates diabetes-related cardiovascular disease, independently of effects on glycaemia [88, 89]. Consequently, it will be interesting to study the role of copper binding in the cardioprotective effects of metformin itself.
Abbreviations
- AMPK:
-
5' AMP-activated protein kinase
- CRTC2:
-
cAMP response element-binding protein-regulated transcription coactivator 2
- MATE:
-
Multidrug and toxic compound extrusion
- OCT:
-
Organic cation transporter
- PKA:
-
Protein kinase A
References
Nathan DM, Buse JB, Davidson MB et al (2009) Medical management of hyperglycaemia in type 2 diabetes mellitus: a consensus algorithm for the initiation and adjustment of therapy. A consensus statement from the American Diabetes Association and the European Association for the Study of Diabetes. Diabetologia 52:17–30
Rodbard H, Jellinger P, Davidson J et al (2009) Statement by an American Association of Clinical Endocrinologists/American College of Endocrinology consensus panel on type 2 diabetes mellitus: an algorithm for glycemic control. Endocr Pract 15:540–559
Inzucchi SE, Bergenstal RM, Buse JB et al (2012) Management of hyperglycaemia in type 2 diabetes: a patient-centered approach. Position statement of the American Diabetes Association (ADA) and the European Association for the Study of Diabetes (EASD). Diabetologia 55:1577–1596
UK Prospective Diabetes Study (UKPDS) Group (1998) Effect of intensive blood-glucose control with metformin on complications in overweight patients with type 2 diabetes (UKPDS 34). Lancet 352:854–865
Libby G, Donnelly LA, Donnan PT, Alessi DR, Morris AD, Evans JM (2009) New users of metformin are at low risk of incident cancer: a cohort study among people with type 2 diabetes. Diabetes Care 32:1620–1625
Evans JMM, Donnelly LA, Emslie-Smith AM, Alessi DR, Morris AD (2005) Metformin and reduced risk of cancer in diabetic patients. BMJ 330:1304–1305
Noto H, Goto A, Tsujimoto T, Noda M (2012) Cancer risk in diabetic patients treated with metformin: a systematic review and meta-analysis. PLoS One 7:e33411
van Staa TP, Patel D, Gallagher AM, de Bruin ML (2012) Glucose-lowering agents and the patterns of risk for cancer: a study with the General Practice Research Database and secondary care data. Diabetologia 55:654–665
Watanabe CK (1918) Studies in the metabolism changes induced by administration of guanidine bases. J Biol Chem 33:253–265
Frank E, Nothmann M, Wagner A (1926) Uber synthetisch dargestellte Körper mit insulinartiger Wirkung auf den normalen und diabetischen Organismus. Klin Wochenschr 5:2100–2107 [article in German]
Bischoff F, Sahyun M, Long ML (1929) Guanidine structure and hypoglycemia. J Biol Chem 81:325–349
Blatherwick NR, Sahyun M, Hill E (1927) Some effects of synthalin on metabolism. J Biol Chem 75:671–683
Luft D, Schilling RM, Eggstein M (1978) Lactic acidosis in biguanide-treated diabetics. Diabetologia 14:75–87
Bodo R, Marks HP (1928) The relation of synthalin to carbohydrate metabolism. J Physiol 65:83–99
Chance B, Hollunger G (1963) Inhibition of electron and energy transfer in mitochondria. J Biol Chem 238:432–438
Pressman BC (1963) The effects of guanidine and alkylguanidines on the energy transfer reactions of mitochondria. J Biol Chem 238:401–409
Davidoff F (1971) Effects of guanidine derivatives on mitochondrial function. J Biol Chem 246:4017–4027
Davidoff F (1968) Effects of guanidine derivatives on mitochondrial function: I. Phenethylbiguanide inhibition of respiration in mitochondria from guinea pig and rat tissues. J Clin Invest 47:2331–2343
Schafer G (1981) Guanidines and biguanides. In: Erecinska M, Wilson DF (eds) Inhibitors of mitochondrial functions. Pergamon Press, New York, pp 165–185
Schafer G (1976) On the mechanism of action of hypoglycemia-producing biguanides, a reevaluation and a molecular theory. Biochem Pharm 25:2005–2014
Davidoff F (1973) Guanidine derivatives in medicine. N Engl J Med 289:141–146
Davidoff F, Carr S (1972) Calcium-like action of phenethylbiguanide and related compounds: inhibition of pyruvate kinase. Proc Natl Acad Sci 69:1957–1961
Logie L, Harthill J, Patel K et al (2012) Cellular responses to the metal-binding properties of metformin. Diabetes 61:1423–1433
Sen D (1969) Ultraviolet spectral studies on metal-biguanide complexes. J Chem Soc A, 2900–2903
Zhu M, Lu L, Yang P, ** X (2002) Bis(1,1-dimethylbiguanido)copper(II) octahydrate. Acta Cryst E58:m217–m219
Ray RK, Kauffman GB (1999) Metal and non-metal biguanide complexes. New Age International Publishers, New Delhi
Ray RK, Kauffman GB (1990) An EPR Study of some copper (II) coordination compounds of substituted biguanides. Part IV. Inorg Chim Acta 174:257–262
Ray RK, Kauffman GB (1990) An EPR study of copper (II)-substituted biguanide complexes. Part III. Inorg Chim Acta 174:237–244
Meyer F, Ipaktchi M, Clauser H (1967) Specific inhibition of gluconeogenesis by biguanides. Nature 213:203–204
Cook DE, Blair JB, Lardy HA (1973) Mode of action of hypoglycemic agents. J Biol Chem 248:5272–5277
Owen MR, Doran E, Halestrap AP (2000) Evidence that metformin exerts its anti-diabetic effects through inhibition of complex 1 of the mitochondrial respiratory chain. Biochem J 348:607–614
El-Mir MY, Nogueira V, Fontaine E, Averet N, Rigoulet M, Leverve X (2000) Dimethylbiguanide inhibits cell respiration via an indirect effect targeted on the respiratory chain complex I. J Biol Chem 275:223–228
Hawley SA, Ross FA, Chevtzoff C et al (2010) Use of cells expressing gamma subunit variants to identify diverse mechanisms of AMPK activation. Cell Metabolism 11:554–565
Pryor HJ, Smyth JE, Quinlan PT, Halestrap AP (1987) Evidence that the flux control coefficient of the respiratory chain is high during gluconeogenesis from lactate in hepatocytes from starved rats. Biochem J 247:449–457
Salpeter SR, Greyber E, Pasternak GA, Salpeter EE (2003) Risk of fatal and nonfatal lactic acidosis with metformin use in type 2 diabetes mellitus: systematic review and meta-analysis. Arch Intern Med 163:2594–2602
Salpeter SR, Greyber E, Pasternak GA, Salpeter EE (2010) Risk of fatal and nonfatal lactic acidosis with metformin use in type 2 diabetes mellitus. Cochrane Database Syst Rev, Issue 4. Art. no.: CD002967. doi:10.1002/14651858.CD002967.pub4
Salpeter S, Greyber E, Pasternak G, Salpeter E (2006) Risk of fatal and nonfatal lactic acidosis with metformin use in type 2 diabetes mellitus. Cochrane Database Syst Rev, Issue 1. Art. no.: CD002967. doi:10.1002/14651858.CD002967.pub2
Brunmair B, Staniek K, Gras F et al (2004) Thiazolidinediones, like metformin, inhibit respiratory complex I: a common mechanism contributing to their antidiabetic actions? Diabetes 53:1052–1059
Zhou G, Myers R, Li Y et al (2001) Role of AMP-activated protein kinase in mechanism of metformin action. J Clin Invest 108:1167–1174
Hardie DG, Ross FA, Hawley SA (2012) AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol 13:251–262
Kahn BB, Alquier T, Carling D, Hardie DG (2005) AMP-activated protein kinase: ancient energy gauge provides clues to modern understanding of metabolism. Cell Metabolism 1:15–25
Perriello G, Misericordia P, Volpi E et al (1994) Acute antihyperglycemic mechanisms of metformin in NIDDM. Evidence for suppression of lipid oxidation and hepatic glucose production. Diabetes 43:920–928
Bain J, Plater L, Elliott M et al (2007) The selectivity of protein kinase inhibitors; a further update. Biochem J 408:297–315
Vogt J, Traynor R, Sapkota GP (2011) The specificities of small molecule inhibitors of the TGFβ and BMP pathways. Cell Signal 23:1831–1842
Shaw RJ, Lamia KA, Vasquez D et al (2005) The kinase LKB1 mediates glucose homeostasis in liver and therapeutic effects of metformin. Science 310:1642–1646
Altarejos JY, Montminy M (2011) CREB and the CRTC co-activators: sensors for hormonal and metabolic signals. Nat Rev Mol Cell Biol 12:141–151
Lin HV, Accili D (2011) Hormonal regulation of hepatic glucose production in health and disease. Cell Metab 14:9–19
Foretz M, Hébrard S, Leclerc J et al (2010) Metformin inhibits hepatic gluconeogenesis in mice independently of the LKB1/AMPK pathway via a decrease in hepatic energy state. J Clin Invest 120:2355–2369
Alessi DR, Sakamoto K, Bayascas JR (2006) LKB1-dependent signaling pathways. Annu Rev Biochem 75:137–163
Koepsell H, Lips K, Volk C (2007) Polyspecific organic cation transporters: structure, function, physiological roles, and biopharmaceutical implications. Pharm Res 24:1227–1251
Rena G, Pearson ER, Sakamoto K (2012) Molecular action and pharmacogenetics of metformin: current understanding of an old drug. Diabetes Management 2:439–452
Shu Y, Sheardown SA, Brown C et al (2007) Effect of genetic variation in the organic cation transporter 1 (OCT1) on metformin action. J Clin Invest 117:1422–1431
Bailey CJ, Puah JA (1986) Effect of metformin on glucose metabolism in mouse soleus muscle. Diabetes Metab 12:212–218
Turban S, Stretton C, Drouin O et al (2012) Defining the contribution of AMPK and PKCs in the regulation of glucose uptake by metformin in skeletal muscle cells. J Biol Chem 287:20088–20099
Hundal HS, Ramlal T, Reyes R, Leiter LA, Klip A (1992) Cellular mechanism of metformin action involves glucose transporter translocation from an intracellular pool to the plasma membrane in L6 muscle cells. Endocrinology 131:1165–1173
Musi N, Hirshman MF, Nygren J et al (2002) Metformin increases AMP-activated protein kinase activity in skeletal muscle of subjects with type 2 diabetes. Diabetes 51:2074–2081
Bailey CJ, Turner RC (1996) Metformin. N Engl J Med 334:574–579
Hardie DG (2007) AMP-activated protein kinase as a drug target. Annu Rev Pharmacol Toxicol 47:185–210
Wilcock C, Bailey CJ (1994) Accumulation of metformin by tissues of the normal and diabetic mouse. Xenobiotica 24:49–57
Morrow VA, Foufelle F, Connell JMC, Petrie JR, Gould GW, Salt IP (2003) Direct activation of AMP-activated protein kinase stimulates nitric-oxide synthesis in human aortic endothelial cells. J Biol Chem 278:31629–31639
Davis BJ, **e Z, Viollet B, Zou MH (2006) Activation of the AMP-activated kinase by antidiabetes drug metformin stimulates nitric oxide synthesis in vivo by promoting the association of heat shock protein 90 and endothelial nitric oxide synthase. Diabetes 55:496–505
Ouslimani N, Peynet J, Bonnefont-Rousselot D, Thérond P, Legrand A, Beaudeux J-L (2005) Metformin decreases intracellular production of reactive oxygen species in aortic endothelial cells. Metabolism 54:829–834
Sanders MJ, Ali ZS, Hegarty BD, Heath R, Snowden MA, Carling D (2007) Defining the mechanism of activation of AMP-activated protein kinase by the small molecule A-769662, a member of the thienopyridone family. J Biol Chem 282:32539–32548
Scott JW, van Denderen BJ, Jorgensen SB et al (2008) Thienopyridone drugs are selective activators of AMP-activated protein kinase β1-containing complexes. Chem Biol 15:1220–1230
Miller RA, Birnbaum MJ (2010) An energetic tale of AMPK-independent effects of metformin. J Clin Invest 120:2267–2270
McGrane MM, El-Maghrabi MR, Pilkis SJ (1983) The interaction of fructose 2,6-bisphosphate and AMP with rat hepatic fructose 1,6-bisphosphatase. J Biol Chem 258:10445–10454
Miller RA, Chu Q, **e J, Foretz M, Viollet B, Birnbaum MJ (2013) Biguanides suppress hepatic glucagon signalling by decreasing production of cyclic AMP. Nature 494:256–260
Gawler DJ, Wilson A, Houslay MD (1989) Metformin treatment of lean and obese Zucker rats modulates the ability of glucagon and insulin to regulate hepatocyte adenylate cyclase activity. J Endocrinol 122:207–212
Alengrin F, Grossi G, Canivet B, Dolais-Kitabgi J (1987) Inhibitory effects of metformin on insulin and glucagon action in rat hepatocytes involve postreceptor alterations. Diabet Metab 13:591–597
Yu B, Pugazhenthi S, Khandelwal RL (1994) Effects of metformin on glucose and glucagon regulated gluconeogenesis in cultured normal and diabetic hepatocytes. Biochem Pharmacol 48:949–954
Houslay MD, Milligan G (1997) Tailoring cAMP-signalling responses through isoform multiplicity. Trends Biochem Sci 22:217–224
Gelling RW, Du XQ, Dichmann DS et al (2003) Lower blood glucose, hyperglucagonemia, and pancreatic α cell hyperplasia in glucagon receptor knockout mice. Proc Natl Acad Sci 100:1438–1443
Kazda CM, Garhyan P, Kelly RP et al (2012) 12-week treatment with glucagon receptor antagonist LY2409021 significantly lowers HbA1c and is well tolerated in patients with type 2 diabetes mellitus. Diabetologia 55(Suppl 1):S51, Abstr. 112
Ouhilal S, Vuguin P, Cui L et al (2012) Hypoglycemia, hyperglucagonemia, and fetoplacental defects in glucagon receptor knockout mice: a role for glucagon action in pregnancy maintenance. Am J Physiol Endocrinol Metab 302:E522–E531
Wulffele MG, Kooy A, de Zeeuw D, Stehouwer CD, Gansevoort RT (2004) The effect of metformin on blood pressure, plasma cholesterol and triglycerides in type 2 diabetes mellitus: a systematic review. J Intern Med 256:1–14
Winegrad AI, Davidson JK, Ricketts HT et al (1971) The University Group Diabetes Program Study pertaining to phenformin. JAMA 217:817
Nattrass M, Alberti KGMM (1978) Biguanides. Diabetologia 14:71–74
Viollet B, Guigas B, Sanz Garcia N, Leclerc J, Foretz M, Andreelli F (2012) Cellular and molecular mechanisms of metformin: an overview. Clin Sci 122:253–270
Shu Y, Brown C, Castro RA et al (2008) Effect of genetic variation in the organic cation transporter 1, OCT1, on metformin pharmacokinetics. Clin Pharmacol Ther 83:273–280
Christensen MMH, Brasch-Andersen C, Green H et al (2011) The pharmacogenetics of metformin and its impact on plasma metformin steady-state levels and glycosylated hemoglobin A1c. Pharmacogenetics and Genomics 21:837–850
Zhou K, Donnelly LA, Kimber CH et al (2009) Reduced-function SLC22A1 polymorphisms encoding organic cation transporter 1 and glycemic response to metformin: a GoDARTS study. Diabetes 58:1434–1439
Van Leeuwen N, Nijpels G, Becker ML et al (2012) A gene variant near ATM is significantly associated with metformin treatment response in type 2 diabetes: a replication and meta-analysis of five cohorts. Diabetologia 55:1971–1977
The GoDARTS and UKPDS Diabetes Pharmacogenetics Study Group & The Wellcome Trust Case Control Consortium 2 (2011) Common variants near ATM are associated with glycemic response to metformin in type 2 diabetes. Common variants near ATM are associated with glycemic response to metformin in type 2 diabetes. Nat Genet 43:117–120
Donnelly LA, Morris AD, Pearson ER (2009) Adherence in patients transferred from immediate release metformin to a sustained release formulation: a population-based study. Diabetes Obes Metab 11:338–342
Christensen M, Bagger JI, Vilsboll T, Knop FK (2011) The alpha-cell as target for type 2 diabetes therapy. The review of diabetic studies: Rev Diabet Stud 8:369–381
Beshgetoor D, Hambidge M (1998) Clinical conditions altering copper metabolism in humans. Am J Clin Nutr 67:1017S–1021S
Cooper GJS, Chan YK, Dissanayake AM et al (2005) Demonstration of a hyperglycemia-driven pathogenic abnormality of copper homeostasis in diabetes and its reversibility by selective chelation: quantitative comparisons between the biology of copper and eight other nutritionally essential elements in normal and diabetic individuals. Diabetes 54:1468–1476
Cooper GJS, Phillips ARJ, Choong SY et al (2004) Regeneration of the heart in diabetes by selective copper chelation. Diabetes 53:2501–2508
Cooper G, Young A, Gamble G et al (2009) A copper(II)-selective chelator ameliorates left-ventricular hypertrophy in type 2 diabetic patients: a randomised placebo-controlled study. Diabetologia 52:715–722
Funding
Graham Rena gratefully acknowledges recent and current support from Diabetes UK, MRC, Tenovus Scotland and The Rank Prize Funds
Duality of interest
The authors declare that there is no duality of interest associated with this manuscript.
Contribution statement
GR, ERP and KS all contributed to the conception, drafting and revision of the manuscript. All authors gave final approval.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Rena, G., Pearson, E.R. & Sakamoto, K. Molecular mechanism of action of metformin: old or new insights?. Diabetologia 56, 1898–1906 (2013). https://doi.org/10.1007/s00125-013-2991-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-013-2991-0