Abstract
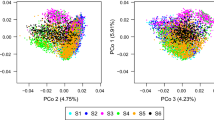
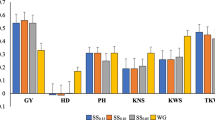
Genomic selection is a promising breeding strategy for rapid improvement of complex traits. The objective of our study was to investigate the prediction accuracy of genomic breeding values through cross validation. The study was based on experimental data of six segregating populations from a half-diallel mating design with 788 testcross progenies from an elite maize breeding program. The plants were intensively phenotyped in multi-location field trials and fingerprinted with 960 SNP markers. We used random regression best linear unbiased prediction in combination with fivefold cross validation. The prediction accuracy across populations was higher for grain moisture (0.90) than for grain yield (0.58). The accuracy of genomic selection realized for grain yield corresponds to the precision of phenoty** at unreplicated field trials in 3–4 locations. As for maize up to three generations are feasible per year, selection gain per unit time is high and, consequently, genomic selection holds great promise for maize breeding programs.



Similar content being viewed by others
References
Bernardo R, Yu J (2007) Prospects for genome-wide selection for quantitative traits in maize. Crop Sci 47:1082–1090
Blanc G, Charcosset A, Mangin B, Gallais A, Moreau L (2006) Connected populations for detecting quantitative trait loci and testing for epistasis: an application in maize. Theor Appl Genet 113:206–224
Calus MPL, Meuwissen THE, de Roos APW, Veerkamp RF (2008) Accuracy of genomic selection using different methods to define haplotypes. Genetics 178:553–561
Carlborg O, Haley CS (2004) Epistasis: too often neglected in complex trait studies? Nat Rev Genet 5:618–625
Crossa J, Campos G, Pérez P, Gianola D, Burgueño J, Araus JL, Makumbi D, Singh RP, Dreisigacker S, Yan J, Arief V, Banziger M, Braun H-J (2010) Prediction of genetic values of quantitative traits in plant breeding using pedigree and molecular markers. Genetics 186:713–724
Daetwyler HD, Pong-Wong R, Villanueva B, Woolliams JA (2010) The impact of genetic architecture on genome-wide evaluation methods. Genetics 185:1021–1031
de Roos APW, Hayes BJ, Goddard ME (2009) Reliability of genomic predictions across multiple populations. Genetics 183:1545–1553
Dekkers JCM (2007) Prediction of response to marker-assisted and genomic selection using selection index theory. J Anim Breed Genet 124:331–341
Habier D, Fernando RL, Dekkers JCM (2007) The impact of genetic relationship information on genome-assisted breeding values. Genetics 177:2389–2397
Hayes BJ, Bowman PJ, Chamberlain AJ, Goddard ME (2009) Invited review: genomic selection in dairy cattle: progress and challenges. J Dairy Sci 92:433–443
Heffner EL, Sorrells ME, Jannink JL (2009) Genomic selection for crop improvement. Crop Sci 49:1–12
Heffner EL, Lorenz AJ, Jannink J-L, Sorrells ME (2010) Plant breeding with genomic selection: gain per unit time and cost. Crop Sci 50:1681–1690
Henderson CR (1984) Application of linear models in animal breeding. University of Guelph, Guelph
Holm S (1979) A simple sequentially rejective multiple test procedure. Scand J Stat 6:65–70
Jannink J-L, Lorenz AJ, Iwata H (2010) Genomic selection in plant breeding: from theory to practice. Brief Funct Genomics 9:166–177
Lande R, Thompson R (1990) Efficiency of marker-assisted selection in the improvement of quantitative traits. Genetics 124:743–756
Liu W, Gowda M, Steinhoff J, Maurer HP, Würschum T, Longin CFH, Cossic F, Reif JC (2011) Association map** in an elite maize breeding population. Theor Appl Genet 123:847–858
Lorenzana R, Bernardo R (2009) Accuracy of genotypic value predictions for marker-based selection in biparental plant populations. Theor Appl Genet 120:151–161
Luan T, Woolliams JA, Lien S, Kent M, Svendsen M, Meuwissen TH (2009) The accuracy of genomic selection in Norwegian red cattle assessed by cross-validation. Genetics 183:1119–1126
Melchinger AE, Utz HF, Schön CC (1998) Quantitative trait locus (QTL) map** using different testers and independent population samples in maize reveals low power of QTL detection and larger bias in estimates of QTL effects. Genetics 149:383–403
Melchinger AE, Utz HF, Piepho HP, Zeng ZB, Schön CC (2007) The role of epistasis in the manifestation of heterosis: a systems-oriented approach. Genetics 177:1815–1825
Meuwissen THE, Goddard ME (2010) Accurate prediction of genetic values for complex traits by whole-genome resequencing. Genetics 185:623–631
Meuwissen THE, Hayes BJ, Goddard ME (2001) Prediction of total genetic value using genome-wide dense marker maps. Genetics 157:1819–1829
Piepho HP (2009) Ridge regression and extensions for genome-wide selection in maize. Crop Sci 49:1165–1176
Reif JC, Liu W, Gowda M, Maurer HP, Möhring J, Fischer S, Schechert A, Würschum T (2010) Genetic basis of agronomically important traits in sugar beet (Beta vulgaris L.) investigated with joint linkage association map**. Theor Appl Genet 121:1489–1499
Steinhoff J, Liu W, Maurer HP, Würschum T, Longin CFH, Reif JC (2011) Variation in allele substitution effects determined with multiple-line QTL-map** in maize. Crop Sci. doi:10.2135/cropsci2011.03.0181
Ter Braak CJF, Boer M, Bink M (2005) Extending Xu’s Bayesian model for estimating polygenic effects using markers of the entire genome. Genetics 170:1435–1438
Van Inghelandt D, Reif JC, Dhillon BS, Flament P, Melchinger AE (2011) Extent and genome-wide distribution of linkage disequilibrium in commercial maize germplasm. Theor Appl Genet 123:11–20
Wegenast T, Longin CFH, Utz HF, Melchinger AE, Maurer HP, Reif JC (2008) Hybrid maize breeding with doubled haploids IV. Number versus size of crosses and importance of parental selection in two-stage selection for testcross performance. Theor Appl Genet 117:251–260
Whittaker JC, Thompson R, Denham MC (2000) Marker-assisted selection using ridge regression. Genet Res 75:249–252
Xu S (2003) Estimating polygenic effects using markers of the entire genome. Genetics 163:789–801
Zhong SQ, Dekkers JCM, Fernando RL, Jannink JL (2009) Factors affecting accuracy from genomic selection in populations derived from multiple inbred lines: a barley case study. Genetics 182:355–364
Acknowledgments
This research was conducted within the Biometric and Bioinformatic Tools for Genomics based Plant Breeding project supported by the German Federal Ministry of Education and Research (BMBF) within the framework of GABI–FUTURE initiative. We greatly appreciate the helpful comments and suggestions of two anonymous reviewers.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Charcosset.
Rights and permissions
About this article
Cite this article
Zhao, Y., Gowda, M., Liu, W. et al. Accuracy of genomic selection in European maize elite breeding populations. Theor Appl Genet 124, 769–776 (2012). https://doi.org/10.1007/s00122-011-1745-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-011-1745-y