Abstract
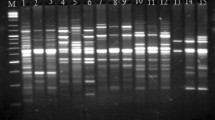
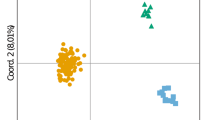
We investigated the use of the polymerase chain reaction (PCR) and the associated random amplification of polymorphic DNA (RAPD) technique to study variation in samples of the sea bass, Dicentrarchus labrax, before and after acclimation to freshwater. Acclimation trials were repeated twice, in 1989 and 1990, for two samples originating from the same broodstock (individuals from different localities along the Italian coasts), with overall mortality rates averaging 94 and 75% in the 2 yr, respectively. Analyses are based on 126 polymorphic RAPD markers, scored in at least 39 individuals for each of the starting and acclimated samples in both years. Analysis of RAPD patterns revealed high levels of DNA polymorphism in both 1989 and 1990 starting samples. The acclimated samples maintained similar polymorphism levels. Shifts in marker frequencies between starting and acclimated samples occurred in both years. A correspondence analysis carried out on multimarker individual profiles suggests that the two starting samples resulted from uneven sampling of the same heterogenous broodstock. This analysis clearly separates RAPD phenotypes from starting and acclimated samples in both years and identifies the RAPD markers responsible for such displacement. Patterns of RAPD variation are compared with previous allozymic studies carried out on the same samples. A major difference between the two studies was the number of markers available. Fewer allozyme loci were studied than were RAPD markers. The cause of repeated shifts for allozyme alleles in replicate experiments were almost certainly due to selection, while statistical chance could explain the repeated shift of only one out of more than 100 RAPD markers. We have shown that RAPD analysis, if carried out carefully, is quite reproducible and sensitive enough to reveal high levels of variation among individuals from the same broodstock. A major drawback of this approach is the still unclear inheritance patterns of RAPD polymorphisms. The use of multivariate analyses is suggested as a possible alternative to traditional population genetics techniques to analyze patterns of variation in the absence of a precise genetic interpretation.
Similar content being viewed by others
References
Allegrucci G, Fortunato C, Cataudella S, Sbordoni V (1994) Acclimation to freshwater of the sea bass: evidence of selective mortality of allozyme genotypes. In: Beaumont AR (ed) Genetic and evolution of aquatic organisms. Chapman and Hall, London, pp 486–502
Arnold LM, Buckner CM, Robinson JJ (1991) Pollen-mediated introgression and hybrid speciation in Louisiana irises. Proc natn Acad Sci USA 88:1398–1402
Benzecri' JP et al. (1973). L'analyse des donnes. II. L'analyse des correspondenses. Dunod, Paris
Black IV WC, DuTeau NM, Puterka GJ, Nechols JR, Pettorini JM (1992) Use of the RAPD polymerase chain reaction to detect DNA polymorphisms in aphids (Homoptera: Aphididae). Bull ent Res 82:151–159
Caccone A, Amato GD, Powell JR (1987) Intraspecific DNA divergence in Drosophila: a study on parthenogenetic D. mercatorum. J molec Biol Evolut 4:343–350
Caswell-Chen EP, Williamson VM, Wu FF (1992) RAPD analysis of Heterodera cruciferae and H. schachtii populations. J Nematol 24:343–351
Clark AG, Lanigan CMS (1993) Prospects for estimating nucleotide divergence with RAPDs. J molec Biol Evolut 10:1096–1111
Ellsworth DL, Rittenhouse KD, Honeycutt RL (1993) Artifactual variation in Randomly Amplified Polymorphic DNA (RAPD) banding patterns. Bio Techniques 14:214–217
Gini C (1912) Variabilità e mutuabilità. Contributo allo studio delle distribuzione e delle relazioni statistiche. Studi Econ. Giur. Fac. Giurisp. R. Univ. Cagliari. Anno III, parte II. P. Cuppini, Bologna, pp 3–158
Hill MO (1973) Reciprocal averaging: an eigenvector method of ordination. J Ecol 61:237–249
Holm S (1979) A simple sequentially rejective multiple test procedure. Scand J Stat 6:65–70
Hu J, Quiros CF (1991) Identification of broccoli and cauliflower cultivars with RAPD markers. Plant Cell Rep 10:505–511
Kambhampati S, Black IV WC, Rai KS (1992) Random amplified polymorphic DNA of mosquito species and populations (Diptera: Culicidae): techniques, statistical analysis and applications. J Med Ent 29: 939–945
Kirpichnikov VS (1981) Genetic basis of fish selection. Springer-Verlag, Berlin
Kresovich S, Williams JGK, McFerson JR, Routman EJ, Schaal BA (1992) Characterization of genetic identities and relationships of Brassica oleracea L. via random amplified polymorphic DNA assay. Theor appl Genet 85:190–196
Lynch M, Milligan BG (1994) Analysis of population genetic structure with RAPD markers. Molec Ecol 3:91–99
Nevo E (1978) Genetic variation in natural populations: patterns and theory. Theor Popul Biol 13:121–177
Rice WR (1989) Analyzing tables of statistical tests. Evolution 43:223–225
Sbordoni V, De Matthaeis E, Cobolli Sbordoni M, La Rosa G, Mattoccia M (1986) Bottleneck effects and the depression of genetic variability in hatchery stocks of Penaeus japonicus (Crustacea, Decapoda). Aquaculture, Amsterdam 57:239–251
Sbordoni V, La Rosa G, Mattoccia M, Cobolli Sbordoni M, De Matthaeis E (1987) Genetic changes in seven generations of hatchery stocks of the kuruma prawn, Penaeus japonicus (Crustacea, Decapoda). In: Trewsk K (ed) Proc World Symp on Selection, Hybridization and Genetic Engineering in Aquaculture, Bordeaux 27–30 May, 1986, Vol I. Bundesforschungsanstalt für Fischerei Berlin, pp 143–155
Tortonese E (1986a) Serranidae (including Anthiidae of some recent authors). In: Whitehead PJP, Bauchot ML, Hureau JC, Nielsen J, Tortonese E (eds) Fishes of the north-eastern Atlantic and the Mediterranean, Vol. 2. Unesco, Paris
Tortonese E (1986b) Moronidae (included in Serranidae by earlier authors, and in Percichthydae sensu Gosline, 1966 by some recent authors). In: Whitehead PJP, Bauchot ML, Hureau JC, Nielsen J, Tortonese E (eds) Fishes of north-eastern Atlantic and the Mediterranean, Vol. 2, Unesco, Paris
Welsh J, Pretzman C, Postic D, Saint Girons I, Baranton G, McClelland M (1992) Genomic fingerprinting by arbitrarily primed polymerase chain reactions resolves Borrelia burgdorferi into three distinct phyletic groups. Int J system Bact 42:370–377
Williams JGK, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV (1990) DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res 18:6531–6535
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Allegrucci, G., Caccone, A., Cataudella, S. et al. Acclimation of the European sea bass to freshwater: monitoring genetic changes by RAPD polymerase chain reaction to detect DNA polymorphisms. Marine Biology 121, 591–599 (1995). https://doi.org/10.1007/BF00349294
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00349294