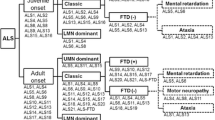
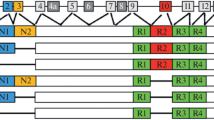
Abstract
Genetic risk factors play a major role in the susceptibility to amyotrophic lateral sclerosis (ALS), the most common motor neuron disease of the adult. Although genetic studies have partially elucidated the genetic background of the disease, a large part of ALS heritability is still missing. In this chapter, we discuss the major genes implicated in the pathogenesis of motor neuron diseases; the clinical, pathological, and genetic links between ALS and frontotemporal dementia; and the vast genotypic and phenotypic heterogeneity of ALS. Lastly, we review the most recent strategies for identification of novel genetic risk factors in ALS, detailing their advantages and potential pitfalls.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Chiò A, Logroscino G, Traynor BJ, et al. Global epidemiology of amyotrophic lateral sclerosis: a systematic review of the published literature. Neuroepidemiology. 2013;41:118–30. https://doi.org/10.1159/000351153.
Alonso A, Logroscino G, Jick SS, Hernán MA. Incidence and lifetime risk of motor neuron disease in the United Kingdom: a population-based study. Eur J Neurol. 2009;16:745–51.
Logroscino G, Traynor BJ, Hardiman O, et al. Descriptive epidemiology of amyotrophic lateral sclerosis: new evidence and unsolved issues. J Neurol Neurosurg Psychiatry. 2008;79:6–11. https://doi.org/10.1136/jnnp.2006.104828.
Statland JM, Barohn RJ, McVey AL, et al. Patterns of weakness, classification of motor neuron disease, and clinical diagnosis of sporadic amyotrophic lateral sclerosis. Neurol Clin. 2015;33:735–48. https://doi.org/10.1016/j.ncl.2015.07.006.
Chiò A, Calvo A, Moglia C, et al. Phenotypic heterogeneity of amyotrophic lateral sclerosis: a population based study. J Neurol Neurosurg Psychiatry. 2011;82:740–6. https://doi.org/10.1136/jnnp.2010.235952.
Ferrari R, Kapogiannis D, Huey ED, Momeni P. FTD and ALS: a tale of two diseases. Curr Alzheimer Res. 2011;8:273–94.
Swinnen B, Robberecht W. The phenotypic variability of amyotrophic lateral sclerosis. Nat Rev Neurol. 2014;10:661–70. https://doi.org/10.1038/nrneurol.2014.184.
Kurland LT, Mulder DW. Epidemiologic investigations of amyotrophic lateral sclerosis. 2. Familial aggregations indicative of dominant inheritance. I. Neurology. 1955;5:182–96.
Kurland LT, Mulder DW. Epidemiologic investigations of amyotrophic lateral sclerosis. 2. Familial aggregations indicative of dominant inheritance. II. Neurology. 1955;5:249–68.
Al-Chalabi A, Fang F, Hanby MF, et al. An estimate of amyotrophic lateral sclerosis heritability using twin data. J Neurol Neurosurg Psychiatry. 2010;81:1324–6. https://doi.org/10.1136/jnnp.2010.207464.
Riggs JE. Longitudinal Gompertzian analysis of amyotrophic lateral sclerosis mortality in the U.S., 1977-1986: evidence for an inherently susceptible population subset. Mech Ageing Dev. 1990;55:207–20.
Hanby MF, Scott KM, Scotton W, et al. The risk to relatives of patients with sporadic amyotrophic lateral sclerosis. Brain J Neurol. 2011;134:3454–7. https://doi.org/10.1093/brain/awr248.
Fang F, Kamel F, Lichtenstein P, et al. Familial aggregation of amyotrophic lateral sclerosis. Ann Neurol. 2009;66:94–9. https://doi.org/10.1002/ana.21580.
Andersen PM, Al-Chalabi A. Clinical genetics of amyotrophic lateral sclerosis: what do we really know? Nat Rev Neurol. 2011;7:603–15. https://doi.org/10.1038/nrneurol.2011.150.
Renton AE, Chiò A, Traynor BJ. State of play in amyotrophic lateral sclerosis genetics. Nat Neurosci. 2014;17:17–23. https://doi.org/10.1038/nn.3584.
Al-Chalabi A, Lewis CM. Modelling the effects of penetrance and family size on rates of sporadic and familial disease. Hum Hered. 2011;71:281–8. https://doi.org/10.1159/000330167.
Byrne S, Bede P, Elamin M, et al. Proposed criteria for familial amyotrophic lateral sclerosis. Amyotroph Lateral Scler. 2011;12:157–9. https://doi.org/10.3109/17482968.2010.545420.
Abel O, Powell JF, Andersen PM, Al-Chalabi A. ALSoD: a user-friendly online bioinformatics tool for amyotrophic lateral sclerosis genetics. Hum Mutat. 2012;33:1345–51. https://doi.org/10.1002/humu.22157.
Kenna KP, McLaughlin RL, Hardiman O, Bradley DG. Using reference databases of genetic variation to evaluate the potential pathogenicity of candidate disease variants. Hum Mutat. 2013;34:836–41. https://doi.org/10.1002/humu.22303.
Al-Chalabi A, Jones A, Troakes C, et al. The genetics and neuropathology of amyotrophic lateral sclerosis. Acta Neuropathol. 2012;124:339–52. https://doi.org/10.1007/s00401-012-1022-4.
Johnson JO, Mandrioli J, Benatar M, et al. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2010;68:857–64. https://doi.org/10.1016/j.neuron.2010.11.036.
Conte A, Lattante S, Zollino M, et al. P525L FUS mutation is consistently associated with a severe form of juvenile amyotrophic lateral sclerosis. Neuromuscul Disord. 2012;22:73–5. https://doi.org/10.1016/j.nmd.2011.08.003.
Ticozzi N, Silani V, LeClerc AL, et al. Analysis of FUS gene mutation in familial amyotrophic lateral sclerosis within an Italian cohort. Neurology. 2009;73:1180–5. https://doi.org/10.1212/WNL.0b013e3181bbff05.
Kenna KP, McLaughlin RL, Byrne S, et al. Delineating the genetic heterogeneity of ALS using targeted high-throughput sequencing. J Med Genet. 2013;50:776–83. https://doi.org/10.1136/jmedgenet-2013-101795.
Chiò A, Calvo A, Mazzini L, et al. Extensive genetics of ALS: a population-based study in Italy. Neurology. 2012;79:1983–9. https://doi.org/10.1212/WNL.0b013e3182735d36.
Lattante S, Conte A, Zollino M, et al. Contribution of major amyotrophic lateral sclerosis genes to the etiology of sporadic disease. Neurology. 2012;79:66–72. https://doi.org/10.1212/WNL.0b013e31825dceca.
Hadano S, Hand CK, Osuga H, et al. A gene encoding a putative GTPase regulator is mutated in familial amyotrophic lateral sclerosis 2. Nat Genet. 2001;29:166–73. https://doi.org/10.1038/ng1001-166.
Yang Y, Hentati A, Deng HX, et al. The gene encoding alsin, a protein with three guanine-nucleotide exchange factor domains, is mutated in a form of recessive amyotrophic lateral sclerosis. Nat Genet. 2001;29:160–5. https://doi.org/10.1038/ng1001-160.
Yeh T-H, Lai S-C, Weng Y-H, et al. Screening for C9orf72 repeat expansions in parkinsonian syndromes. Neurobiol Aging. 2013;34:1311.e3–4. https://doi.org/10.1016/j.neurobiolaging.2012.09.002.
Orlacchio A, Babalini C, Borreca A, et al. SPATACSIN mutations cause autosomal recessive juvenile amyotrophic lateral sclerosis. Brain J Neurol. 2010;133:591–8. https://doi.org/10.1093/brain/awp325.
Nishimura AL, Mitne-Neto M, Silva HCA, et al. A mutation in the vesicle-trafficking protein VAPB causes late-onset spinal muscular atrophy and amyotrophic lateral sclerosis. Am J Hum Genet. 2004;75:822–31. https://doi.org/10.1086/425287.
Greenway MJ, Andersen PM, Russ C, et al. ANG mutations segregate with familial and “sporadic” amyotrophic lateral sclerosis. Nat Genet. 2006;38:411–3. https://doi.org/10.1038/ng1742.
Chow CY, Landers JE, Bergren SK, et al. Deleterious variants of FIG4, a phosphoinositide phosphatase, in patients with ALS. Am J Hum Genet. 2009;84:85–8. https://doi.org/10.1016/j.ajhg.2008.12.010.
Maruyama H, Morino H, Ito H, et al. Mutations of optineurin in amyotrophic lateral sclerosis. Nature. 2010;465:223–6. https://doi.org/10.1038/nature08971.
Rosen DR, Siddique T, Patterson D, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. https://doi.org/10.1038/362059a0.
McCord JM, Fridovich I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein). J Biol Chem. 1969;244:6049–55.
Andersen PM, Sims KB, **n WW, et al. Sixteen novel mutations in the Cu/Zn superoxide dismutase gene in amyotrophic lateral sclerosis: a decade of discoveries, defects and disputes. Amyotroph Lateral Scler Mot Neuron Disord. 2003;4:62–73.
Cudkowicz ME, McKenna-Yasek D, Sapp PE, et al. Epidemiology of mutations in superoxide dismutase in amyotrophic lateral sclerosis. Ann Neurol. 1997;41:210–21. https://doi.org/10.1002/ana.410410212.
Juneja T, Pericak-Vance MA, Laing NG, et al. Prognosis in familial amyotrophic lateral sclerosis: progression and survival in patients with glu100gly and ala4val mutations in Cu,Zn superoxide dismutase. Neurology. 1997;48:55–7.
Aoki M, Ogasawara M, Matsubara Y, et al. Mild ALS in Japan associated with novel SOD mutation. Nat Genet. 1993;5:323–4. https://doi.org/10.1038/ng1293-323.
Luigetti M, Madia F, Conte A, et al. SOD1 G93D mutation presenting as paucisymptomatic amyotrophic lateral sclerosis. Amyotroph Lateral Scler. 2009;10:479–82. https://doi.org/10.3109/17482960802302261.
Orrell RW, Habgood JJ, Malaspina A, et al. Clinical characteristics of SOD1 gene mutations in UK families with ALS. J Neurol Sci. 1999;169:56–60.
Andersen PM, Forsgren L, Binzer M, et al. Autosomal recessive adult-onset amyotrophic lateral sclerosis associated with homozygosity for Asp90Ala CuZn-superoxide dismutase mutation. A clinical and genealogical study of 36 patients. Brain J Neurol. 1996;119(Pt 4):1153–72.
Al-Chalabi A, Andersen PM, Chioza B, et al. Recessive amyotrophic lateral sclerosis families with the D90A SOD1 mutation share a common founder: evidence for a linked protective factor. Hum Mol Genet. 1998;7:2045–50.
Neumann M, Sampathu DM, Kwong LK, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–3. https://doi.org/10.1126/science.1134108.
Sreedharan J, Blair IP, Tripathi VB, et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–72. https://doi.org/10.1126/science.1154584.
Van Deerlin VM, Leverenz JB, Bekris LM, et al. TARDBP mutations in amyotrophic lateral sclerosis with TDP-43 neuropathology: a genetic and histopathological analysis. Lancet Neurol. 2008;7:409–16. https://doi.org/10.1016/S1474-4422(08)70071-1.
Kwiatkowski TJ, Bosco DA, Leclerc AL, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–8. https://doi.org/10.1126/science.1166066.
Vance C, Rogelj B, Hortobágyi T, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–11. https://doi.org/10.1126/science.1165942.
Ravits J, Paul P, Jorg C. Focality of upper and lower motor neuron degeneration at the clinical onset of ALS. Neurology. 2007;68:1571–5. https://doi.org/10.1212/01.wnl.0000260965.20021.47.
Gourie-Devi M, Nalini A, Sandhya S. Early or late appearance of “dropped head syndrome” in amyotrophic lateral sclerosis. J Neurol Neurosurg Psychiatry. 2003;74:683–6.
Munoz DG, Neumann M, Kusaka H, et al. FUS pathology in basophilic inclusion body disease. Acta Neuropathol. 2009;118:617–27. https://doi.org/10.1007/s00401-009-0598-9.
Neumann M, Rademakers R, Roeber S, et al. A new subtype of frontotemporal lobar degeneration with FUS pathology. Brain J Neurol. 2009;132:2922–31. https://doi.org/10.1093/brain/awp214.
Seelaar H, Klijnsma KY, de Koning I, et al. Frequency of ubiquitin and FUS-positive, TDP-43-negative frontotemporal lobar degeneration. J Neurol. 2010;257:747–53. https://doi.org/10.1007/s00415-009-5404-z.
Ratti A, Buratti E. Physiological functions and pathobiology of TDP-43 and FUS/TLS proteins. J Neurochem. 2016;138(Suppl 1):95–111. https://doi.org/10.1111/jnc.13625.
Fujii R, Okabe S, Urushido T, et al. The RNA binding protein TLS is translocated to dendritic spines by mGluR5 activation and regulates spine morphology. Curr Biol. 2005;15:587–93. https://doi.org/10.1016/j.cub.2005.01.058.
Wang I-F, Wu L-S, Chang H-Y, Shen C-KJ. TDP-43, the signature protein of FTLD-U, is a neuronal activity-responsive factor. J Neurochem. 2008;105:797–806. https://doi.org/10.1111/j.1471-4159.2007.05190.x.
Liu-Yesucevitz L, Bassell GJ, Gitler AD, et al. Local RNA translation at the synapse and in disease. J Neurosci. 2011;31:16086–93. https://doi.org/10.1523/JNEUROSCI.4105-11.2011.
Fallini C, Bassell GJ, Rossoll W. The ALS disease protein TDP-43 is actively transported in motor neuron axons and regulates axon outgrowth. Hum Mol Genet. 2012;21:3703–18. https://doi.org/10.1093/hmg/dds205.
DeJesus-Hernandez M, Mackenzie IR, Boeve BF, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–56. https://doi.org/10.1016/j.neuron.2011.09.011.
Renton AE, Majounie E, Waite A, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–68. https://doi.org/10.1016/j.neuron.2011.09.010.
Majounie E, Renton AE, Mok K, et al. Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. Lancet Neurol. 2012;11:323–30. https://doi.org/10.1016/S1474-4422(12)70043-1.
Farg MA, Sundaramoorthy V, Sultana JM, et al. C9ORF72, implicated in amyotrophic lateral sclerosis and frontotemporal dementia, regulates endosomal trafficking. Hum Mol Genet. 2014;23:3579–95. https://doi.org/10.1093/hmg/ddu068.
Ji YJ, Ugolino J, Brady NR, et al. Systemic deregulation of autophagy upon loss of ALS- and FTD-linked C9orf72. Autophagy. 2017;13:1254–5. https://doi.org/10.1080/15548627.2017.1299312.
Haeusler AR, Donnelly CJ, Periz G, et al. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature. 2014;507:195–200. https://doi.org/10.1038/nature13124.
Ash PEA, Bieniek KF, Gendron TF, et al. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77:639–46. https://doi.org/10.1016/j.neuron.2013.02.004.
Mori K, Weng S-M, Arzberger T, et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013;339:1335–8. https://doi.org/10.1126/science.1232927.
Byrne S, Elamin M, Bede P, et al. Cognitive and clinical characteristics of patients with amyotrophic lateral sclerosis carrying a C9orf72 repeat expansion: a population-based cohort study. Lancet Neurol. 2012;11:232–40. https://doi.org/10.1016/S1474-4422(12)70014-5.
Gijselinck I, Van Langenhove T, van der Zee J, et al. A C9orf72 promoter repeat expansion in a Flanders-Belgian cohort with disorders of the frontotemporal lobar degeneration-amyotrophic lateral sclerosis spectrum: a gene identification study. Lancet Neurol. 2012;11:54–65. https://doi.org/10.1016/S1474-4422(11)70261-7.
Snowden JS, Adams J, Harris J, et al. Distinct clinical and pathological phenotypes in frontotemporal dementia associated with MAPT, PGRN and C9orf72 mutations. Amyotroph Lateral Scler Frontotemporal Degener. 2015;16:497–505. https://doi.org/10.3109/21678421.2015.1074700.
Simón-Sánchez J, Dopper EGP, Cohn-Hokke PE, et al. The clinical and pathological phenotype of C9ORF72 hexanucleotide repeat expansions. Brain J Neurol. 2012;135:723–35. https://doi.org/10.1093/brain/awr353.
Beck J, Poulter M, Hensman D, et al. Large C9orf72 hexanucleotide repeat expansions are seen in multiple neurodegenerative syndromes and are more frequent than expected in the UK population. Am J Hum Genet. 2013;92:345–53. https://doi.org/10.1016/j.ajhg.2013.01.011.
Majounie E, Abramzon Y, Renton AE, et al. Repeat expansion in C9ORF72 in Alzheimer’s disease. N Engl J Med. 2012;366:283–4. https://doi.org/10.1056/NEJMc1113592.
Lindquist SG, Duno M, Batbayli M, et al. Corticobasal and ataxia syndromes widen the spectrum of C9ORF72 hexanucleotide expansion disease. Clin Genet. 2013;83:279–83. https://doi.org/10.1111/j.1399-0004.2012.01903.x.
Lesage S, Le Ber I, Condroyer C, et al. C9orf72 repeat expansions are a rare genetic cause of parkinsonism. Brain J Neurol. 2013;136:385–91. https://doi.org/10.1093/brain/aws357.
Hensman Moss DJ, Poulter M, Beck J, et al. C9orf72 expansions are the most common genetic cause of Huntington disease phenocopies. Neurology. 2014;82:292–9. https://doi.org/10.1212/WNL.0000000000000061.
Novarino G, Fenstermaker AG, Zaki MS, et al. Exome sequencing links corticospinal motor neuron disease to common neurodegenerative disorders. Science. 2014;343:506–11. https://doi.org/10.1126/science.1247363.
Chen Y-Z, Bennett CL, Huynh HM, et al. DNA/RNA helicase gene mutations in a form of juvenile amyotrophic lateral sclerosis (ALS4). Am J Hum Genet. 2004;74:1128–35. https://doi.org/10.1086/421054.
Deng H-X, Chen W, Hong S-T, et al. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature. 2011;477:211–5. https://doi.org/10.1038/nature10353.
C-H W, Fallini C, Ticozzi N, et al. Mutations in the profilin 1 gene cause familial amyotrophic lateral sclerosis. Nature. 2012;488:499–503. https://doi.org/10.1038/nature11280.
Tennessen JA, Bigham AW, O’Connor TD, et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–9. https://doi.org/10.1126/science.1219240.
Lee S, Abecasis GR, Boehnke M, Lin X. Rare-variant association analysis: study designs and statistical tests. Am J Hum Genet. 2014;95:5–23. https://doi.org/10.1016/j.ajhg.2014.06.009.
Cirulli ET, Lasseigne BN, Petrovski S, et al. Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. Science. 2015;347:1436–41. https://doi.org/10.1126/science.aaa3650.
Smith BN, Ticozzi N, Fallini C, et al. Exome-wide rare variant analysis identifies TUBA4A mutations associated with familial ALS. Neuron. 2014;84:324–31. https://doi.org/10.1016/j.neuron.2014.09.027.
Kenna KP, van Doormaal PTC, Dekker AM, et al. NEK1 variants confer susceptibility to amyotrophic lateral sclerosis. Nat Genet. 2016;48:1037–42. https://doi.org/10.1038/ng.3626.
James PA, Talbot K. The molecular genetics of non-ALS motor neuron diseases. Biochim Biophys Acta. 2006;1762:986–1000. https://doi.org/10.1016/j.bbadis.2006.04.003.
Beetz C, Pieber TR, Hertel N, et al. Exome sequencing identifies a REEP1 mutation involved in distal hereditary motor neuropathy type V. Am J Hum Genet. 2012;91:139–45. https://doi.org/10.1016/j.ajhg.2012.05.007.
Züchner S, Wang G, Tran-Viet K-N, et al. Mutations in the novel mitochondrial protein REEP1 cause hereditary spastic paraplegia type 31. Am J Hum Genet. 2006;79:365–9. https://doi.org/10.1086/505361.
Auer PL, Lettre G. Rare variant association studies: considerations, challenges and opportunities. Genome Med. 2015;7:16. https://doi.org/10.1186/s13073-015-0138-2.
van Rheenen W, Shatunov A, Dekker AM, et al. Genome-wide association analyses identify new risk variants and the genetic architecture of amyotrophic lateral sclerosis. Nat Genet. 2016;48:1043–8. https://doi.org/10.1038/ng.3622.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer International Publishing AG, part of Springer Nature
About this chapter
Cite this chapter
Ticozzi, N., Silani, V. (2018). Genotypic and Phenotypic Heterogeneity in Amyotrophic Lateral Sclerosis. In: Galimberti, D., Scarpini, E. (eds) Neurodegenerative Diseases. Springer, Cham. https://doi.org/10.1007/978-3-319-72938-1_13
Download citation
DOI: https://doi.org/10.1007/978-3-319-72938-1_13
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-72937-4
Online ISBN: 978-3-319-72938-1
eBook Packages: MedicineMedicine (R0)