Abstract
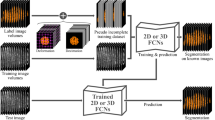
Brain lesion segmentation provides a valuable tool for clinical diagnosis, and convolutional neural networks (CNNs) have achieved unprecedented success in the task. Data augmentation is a widely used strategy that improves the training of CNNs, and the design of the augmentation method for brain lesion segmentation is still an open problem. In this work, we propose a simple data augmentation approach, dubbed as CarveMix, for CNN-based brain lesion segmentation. Like other “mix"-based methods, such as Mixup and CutMix, CarveMix stochastically combines two existing labeled images to generate new labeled samples. Yet, unlike these augmentation strategies based on image combination, CarveMix is lesion-aware, where the combination is performed with an attention on the lesions and a proper annotation is created for the generated image. Specifically, from one labeled image we carve a region of interest (ROI) according to the lesion location and geometry, and the size of the ROI is sampled from a probability distribution. The carved ROI then replaces the corresponding voxels in a second labeled image, and the annotation of the second image is replaced accordingly as well. In this way, we generate new labeled images for network training and the lesion information is preserved. To evaluate the proposed method, experiments were performed on two brain lesion datasets. The results show that our method improves the segmentation accuracy compared with other simple data augmentation approaches.
X. Zhang and C. Liu—Equal contribution.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
Notes
- 1.
Note that CarveMix can also be integrated with other segmentation frameworks if they are shown superior to nnU-net.
References
Barber, P.A., Demchuk, A.M., Zhang, J., Buchan, A.M.: Validity and reliability of a quantitative computed tomography score in predicting outcome of hyperacute stroke before thrombolytic therapy. The Lancet 355(9216), 1670–1674 (2000)
Bdair, T., Wiestler, B., Navab, N., Albarqouni, S.: ROAM: random layer mixup for semi-supervised learning in medical imaging. ar**v preprint ar**v:2003.09439 (2020)
Chaitanya, K., et al.: Semi-supervised task-driven data augmentation for medical image segmentation. Med. Image Anal. 68, 101934 (2021)
Chen, S., Dobriban, E., Lee, J.H.: A group-theoretic framework for data augmentation. J. Mach. Learn. Res. 21, 1–71 (2019)
Çiçek, Ö., Abdulkadir, A., Lienkamp, S.S., Brox, T., Ronneberger, O.: 3D U-Net: learning dense volumetric segmentation from sparse annotation. In: Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W. (eds.) MICCAI 2016. LNCS, vol. 9901, pp. 424–432. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46723-8_49
Gontijo-Lopes, R., Smullin, S., Cubuk, E.D., Dyer, E.: Tradeoffs in data augmentation: an empirical study. In: International Conference on Learning Representations (2021)
Ikram, M.A., et al.: Brain tissue volumes in relation to cognitive function and risk of dementia. Neurobiol. Aging 31(3), 378–386 (2010)
Isensee, F., Jaeger, P.F., Kohl, S.A., Petersen, J., Maier-Hein, K.H.: nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 18(2), 203–211 (2021)
Kamnitsas, K., et al.: Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med. Image Anal. 36, 61–78 (2017)
Li, Z., Kamnitsas, K., Glocker, B.: Analyzing overfitting under class imbalance in neural networks for image segmentation. IEEE Trans. Med. Imaging 40, 1065–1077 (2020)
Liew, S.L., et al.: A large, open source dataset of stroke anatomical brain images and manual lesion segmentations. Sci. Data 5, 180011 (2018)
Pesteie, M., Abolmaesumi, P., Rohling, R.N.: Adaptive augmentation of medical data using independently conditional variational auto-encoders. IEEE Trans. Med. Imaging 38(12), 2807–2820 (2019)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Tajbakhsh, N., Jeyaseelan, L., Li, Q., Chiang, J.N., Wu, Z., Ding, X.: Embracing imperfect datasets: a review of deep learning solutions for medical image segmentation. Med. Image Anal. 63, 10169 (2020)
Yun, S., Han, D., Oh, S.J., Chun, S., Choe, J., Yoo, Y.: CutMix: regularization strategy to train strong classifiers with localizable features. In: Proceedings of the IEEE/CVF International Conference on Computer Vision, pp. 6023–6032 (2019)
Zhang, H., Cisse, M., Dauphin, Y.N., Lopez-Paz, D.: mixup: Beyond empirical risk minimization. In: International Conference on Learning Representations (2018)
Acknowledgement
This work is supported by Bei**g Natural Science Foundation (L192058 & 7192108).
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this paper
Cite this paper
Zhang, X. et al. (2021). CarveMix: A Simple Data Augmentation Method for Brain Lesion Segmentation. In: de Bruijne, M., et al. Medical Image Computing and Computer Assisted Intervention – MICCAI 2021. MICCAI 2021. Lecture Notes in Computer Science(), vol 12901. Springer, Cham. https://doi.org/10.1007/978-3-030-87193-2_19
Download citation
DOI: https://doi.org/10.1007/978-3-030-87193-2_19
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-87192-5
Online ISBN: 978-3-030-87193-2
eBook Packages: Computer ScienceComputer Science (R0)