Abstract
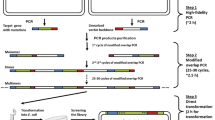
Multi-site-saturation mutagenesis allows altering of “localizable” properties such as activity and selectivity and enables the discovery of cooperative amino acid substitutions which are unlikely to be discovered by saturating single codons individually or iteratively. The herein presented method “OmniChange” does not require any DNA modifying enzyme (e.g., endonucleases or ligases), and diverse mutant libraries with up to five simultaneously saturated positions are generated in a robust and technically simple manner in four steps. The key feature of the OmniChange method is a highly efficient chemical cleavage of phosphorothiolated nucleotides by ethanol-iodine to generate 12-nucleotide-long 5′ overhangs in double-stranded DNA. The generated vector and inserts can be hybridized in a one-pot assembly leading to fully functional mutagenic plasmids, and the employed E. coli host can easily ligate up to 10 DNA nicks without any further enzymatic treatment. OmniChange is furthermore a reliable and general tool for multi-DNA fragment assembly which is DNA sequence independent.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Reetz MT, Carballeira JD, Vogel A (2006) Iterative saturation mutagenesis on the basis of B factors as a strategy for increasing protein thermostability. Angew Chem Int Ed Engl 45:7745–7751
Shivange AV, Marienhagen J, Mundhada H et al (2009) Advances in generating functional diversity for directed protein evolution. Curr Opin Chem Biol 13:19–25
Dennig A, Shivange AV, Marienhagen J, Schwaneberg U (2011) OmniChange: the sequence independent method for simultaneous site-saturation of five codons. PLoS One 6:e26222
Reetz MT, Bocola M, Carballeira JD et al (2005) Expanding the range of substrate acceptance of enzymes: combinatorial active-site saturation test. Angew Chem Int Ed Engl 44:4192–4196
Hogrefe HH, Cline J, Youngblood GL, Allen RM (2002) Creating randomized amino acid libraries with the QuikChange Multi Site-Directed Mutagenesis Kit. Biotechniques 33:1158–1160
Eckstein F, Gish G (1989) Phosphorothioates in molecular biology. Trends Biochem Sci 14:97–100
Whitehouse CJ, Bell SG, Wong LL (2012) P450(BM3) (CYP102A1): connecting the dots. Chem Soc Rev 41:1218–1260
Acknowledgments
The authors acknowledge BASF SE (Ludwigshafen, Germany) for financial support during development of the OmniChange method.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer Science+Business Media New York
About this protocol
Cite this protocol
Dennig, A., Marienhagen, J., Ruff, A.J., Schwaneberg, U. (2014). OmniChange: Simultaneous Site Saturation of Up to Five Codons. In: Gillam, E., Copp, J., Ackerley, D. (eds) Directed Evolution Library Creation. Methods in Molecular Biology, vol 1179. Springer, New York, NY. https://doi.org/10.1007/978-1-4939-1053-3_9
Download citation
DOI: https://doi.org/10.1007/978-1-4939-1053-3_9
Published:
Publisher Name: Springer, New York, NY
Print ISBN: 978-1-4939-1052-6
Online ISBN: 978-1-4939-1053-3
eBook Packages: Springer Protocols