Abstract
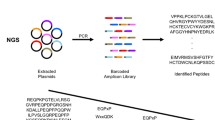
Phage display technique has a multitude of applications such as epitope map**, organ targeting, therapeutic antibody engineering and vaccine design. One area of particular importance is the detection of cancers in early stages, where the discovery of binding motifs and epitopes is critical. While several techniques exist to characterize phages, Next Generation Sequencing (NGS) stands out for its ability to provide detailed insights into antibody binding sites on antigens. However, when dealing with NGS data, identifying regulatory motifs poses significant challenges. Existing methods often lack scalability for large datasets, rely on prior knowledge about the number of motifs, and exhibit low accuracy. In this paper, we present a novel approach for identifying regulatory motifs in NGS data. Our method leverages results from graph theory to overcome the limitations of existing techniques.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Alipanahi, B., Delong, A., Weirauch, M.T., Frey, B.J.: Predicting the sequence specificities of dna-and rna-binding proteins by deep learning. Nat. Biotechnol. 33(8), 831–838 (2015)
Andreatta, M., Lund, O., Nielsen, M.: Simultaneous alignment and clustering of peptide data using a gibbs sampling approach. Bioinformatics 29(1), 8–14 (2013)
Ash, R.B.: Information Theory. Courier Corporation, North Chelmsford (2012)
Bailey, T.L., Elkan, C., et al.: Fitting a mixture model by expectation maximization to discover motifs in bipolymers (1994)
Bratkovič, T.: Progress in phage display: evolution of the technique and its applications. Cell. Mol. Life Sci. 67(5), 749–767 (2010)
Choi, I.G., Kwon, J., Kim, S.H.: Local feature frequency profile: a method to measure structural similarity in proteins. Proc. Natl. Acad. Sci. 101(11), 3797–3802 (2004)
Christiansen, A., et al.: High-throughput sequencing enhanced phage display enables the identification of patient-specific epitope motifs in serum. Sci. Rep. 5(1), 1–13 (2015)
Crooks, G.E., Hon, G., Chandonia, J.M., Brenner, S.E.: Weblogo: a sequence logo generator. Genome Res. 14(6), 1188–1190 (2004)
Dinkel, H., et al.: The eukaryotic linear motif resource elm: 10 years and counting. Nucleic Acids Res. 42(D1), D259–D266 (2014)
Gale, D., Shapley, L.S.: College admissions and the stability of marriage. Am. Math. Mon. 69(1), 9–15 (1962)
Gerasimov, E., Zelikovsky, A., Măndoiu, I., Ionov, Y.: Identification of cancer-specific motifs in mimotope profiles of serum antibody repertoire. BMC Bioinf. 18(8), 1–6 (2017)
Geysen, H.M., Rodda, S.J., Mason, T.J.: A priori delineation of a peptide which mimics a discontinuous antigenic determinant. Mol. Immunol. 23(7), 709–715 (1986)
Gupta, S., Stamatoyannopoulos, J.A., Bailey, T.L., Noble, W.S.: Quantifying similarity between motifs. Genome Biol. 8, 1–9 (2007)
Ionov, Y., Rogovskyy, A.S.: Comparison of motif-based and whole-unique-sequence-based analyses of phage display library datasets generated by biopanning of anti-borrelia burgdorferi immune sera. PLoS ONE 15(1), e0226378 (2020)
Kim, T., et al.: Musi: an integrated system for identifying multiple specificity from very large peptide or nucleic acid data sets. Nucleic Acids Res. 40(6), e47–e47 (2012)
Knittelfelder, R., Riemer, A.B., Jensen-Jarolim, E.: Mimotope vaccination-from allergy to cancer. Expert Opin. Biol. Ther. 9(4), 493–506 (2009)
Kon, M.A., Fan, Y., Holloway, D., DeLisi, C.: Svmotif: a machine learning motif algorithm. In: Sixth International Conference on Machine Learning and Applications (ICMLA 2007), pp. 573–580. IEEE (2007)
Krejci, A., Hupp, T.R., Lexa, M., Vojtesek, B., Muller, P.: Hammock: a hidden markov model-based peptide clustering algorithm to identify protein-interaction consensus motifs in large datasets. Bioinformatics 32(1), 9–16 (2016)
Lawrence, C.E., Altschul, S.F., Boguski, M.S., Liu, J.S., Neuwald, A.F., Wootton, J.C.: Detecting subtle sequence signals: a gibbs sampling strategy for multiple alignment. Science 262(5131), 208–214 (1993)
Liu, X.S., Brutlag, D.L., Liu, J.S.: An algorithm for finding protein-dna binding sites with applications to chromatin-immunoprecipitation microarray experiments. Nat. Biotechnol. 20(8), 835–839 (2002)
Macdougall, I.C., et al.: A peptide-based erythropoietin-receptor agonist for pure red-cell aplasia. N. Engl. J. Med. 361(19), 1848–1855 (2009)
Murphy, K., Weaver, C.: Janeway’s Immunobiology. Garland Science, New York City (2016)
Nielsen, M., Lund, O.: Nn-align. an artificial neural network-based alignment algorithm for mhc class ii peptide binding prediction. BMC Bioinf. 10(1), 296 (2009)
Nielsen, M., et al.: Improved prediction of mhc class i and class ii epitopes using a novel gibbs sampling approach. Bioinformatics 20(9), 1388–1397 (2004)
Pietrokovski, S.: Searching databases of conserved sequence regions by aligning protein multiple-alignments. Nucleic Acids Res. 24(19), 3836–3845 (1996)
Rentero Rebollo, I., Sabisz, M., Baeriswyl, V., Heinis, C.: Identification of target-binding peptide motifs by high-throughput sequencing of phage-selected peptides. Nucleic Acids Res. 42(22), e169 (2014)
Rodi, D.J., Janes, R.W., Sanganee, H.J., Holton, R.A., Wallace, B., Makowski, L.: Screening of a library of phage-displayed peptides identifies human bcl-2 as a taxol-binding protein. J. Mol. Biol. 285(1), 197–203 (1999)
Roepcke, S., Grossmann, S., Rahmann, S., Vingron, M.: T-reg comparator: an analysis tool for the comparison of position weight matrices. Nucleic Acids Res. 33(suppl_2), W438–W441 (2005)
Roth, F.P., Hughes, J.D., Estep, P.W., Church, G.M.: Finding dna regulatory motifs within unaligned noncoding sequences clustered by whole-genome mrna quantitation. Nat. Biotechnol. 16(10), 939–945 (1998)
Schones, D.E., Sumazin, P., Zhang, M.Q.: Similarity of position frequency matrices for transcription factor binding sites. Bioinformatics 21(3), 307–313 (2005)
Smith, G.P.: Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science 228(4705), 1315–1317 (1985)
Smith, G.P., Petrenko, V.A.: Phage display. Chem. Rev. 97(2), 391–410 (1997)
Thom, G., et al.: Probing a protein-protein interaction by in vitro evolution. Proc. Natl. Acad. Sci. 103(20), 7619–7624 (2006)
Tong, A.H.Y., et al.: A combined experimental and computational strategy to define protein interaction networks for peptide recognition modules. Science 295(5553), 321–324 (2002)
Van Regenmortel, M.H.V.: Specificity, polyspecificity and heterospecificity of antibody-antigen recognition. In: HIV/AIDS: Immunochemistry, Reductionism and Vaccine Design, pp. 39–56. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-32459-9_4
Wang, L.F., Yu, M.: Epitope identification and discovery using phage display libraries: applications in vaccine development and diagnostics. Curr. Drug Targets 5(1), 1–15 (2004)
Wang, T., Stormo, G.D.: Combining phylogenetic data with co-regulated genes to identify regulatory motifs. Bioinformatics 19(18), 2369–2380 (2003)
Zhong, L., Coe, S.P., Stromberg, A.J., Khattar, N.H., Jett, J.R., Hirschowitz, E.A.: Profiling tumor-associated antibodies for early detection of non-small cell lung cancer. J. Thorac. Oncol. 1(6), 513–519 (2006)
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Saghaian, H., Skums, P., Ionov, Y., Zelikovsky, A. (2023). Graph-Based Motif Discovery in Mimotope Profiles of Serum Antibody Repertoire. In: Guo, X., Mangul, S., Patterson, M., Zelikovsky, A. (eds) Bioinformatics Research and Applications. ISBRA 2023. Lecture Notes in Computer Science(), vol 14248. Springer, Singapore. https://doi.org/10.1007/978-981-99-7074-2_17
Download citation
DOI: https://doi.org/10.1007/978-981-99-7074-2_17
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-99-7073-5
Online ISBN: 978-981-99-7074-2
eBook Packages: Computer ScienceComputer Science (R0)