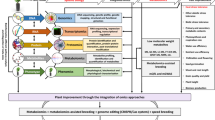
Abstract
Metabolomics is the analysis of micro-biomolecules related to the metabolic processes of a living organism. It has a strong connection between the genotype and phenotype of an organism. Metabolomics is like other “omics” technologies and can handle large-scale, complex, and dynamic databases. As a result, various data processing techniques are needed to retrieve biologically relevant knowledge from a living organism. Those data processing workflows of metabolomics research are typically complicated and require many phases or steps. This chapter will address the application and role of metabolites in the growth, development, and produce chemical compounds for defense against biotic and abiotic stress of rice and other crops. Hence, it is important to know the metabolic process of plants under different environmental conditions. Thus, metabolomic techniques may be used to elucidate the roles of unknown genes by using natural variations and mutations in target plants. These metabolomic methods could be useful in crop breeding, where important plant traits like taste, yield, and grain yield quality are strongly linked to metabolic conditions. Various data analysis techniques are used for metabolomics studies and metabolomics experiments. These methods can identify specific metabolites that help to improve stress resistance and disease resistance. We also discuss the several available computational techniques and tools that can assist in the biological interpretation of metabolomics data. We also introduce emerging methods for designing genome-scale metabolic models to analyze cellular metabolism.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
Abbreviations
- 2D:
-
Two-dimensional
- AAA:
-
Aromatic amino acids
- ADH:
-
Arogenate dehydrogenase
- BL-SOM:
-
Batch-learning self-organizing map
- CE:
-
Capillary electrophoresis
- CE-MS:
-
Capillary electrophoresis-mass spectrometry
- CT:
-
Cold tolerance
- DAD:
-
Diode array detector
- DIMS:
-
Direct infusion mass spectrometry
- FT-ICR:
-
Fourier transform ion cyclotron resonance
- GABA:
-
Gluconeogenesis-aminobutyric acid
- GC-MS:
-
Gas chromatography-mass spectrometry
- GM:
-
Genetically modified
- HPTLC:
-
High-performance thin-layer chromatography
- HRMS:
-
High-resolution mass spectrometry
- LC:
-
Liquid chromatography
- LC-MS:
-
Liquid chromatography-mass spectrometry
- LT:
-
Low-temperature
- MS:
-
Mass-spectrometry
- NMR:
-
Nuclear magnetic resonance spectroscopy
- PCA:
-
Principal component analysis
- PGPR:
-
Plant growth-promoting rhizobacteria
- PLS-DA:
-
Partial least squares discriminant analysis
- SAR:
-
Systemic acquired resistance
- TOF:
-
Time-of-flight
- UPLC:
-
Ultra-performance liquid chromatography
- WRC:
-
World Rice Core
References
FAO. FAO: Rice Market Monitor (RMM). 2018. http://www.fao.org/economic/est/publications/rice-publications/rice-market-monitor-rmm/en/. Accessed 26 Apr 2021.
Kennedy G, Burlingame B. Analysis of food composition data on rice from a plant genetic resources perspective. Food Chem. 2003;80:589–96.
Vlachos A, Arvanitoyannis IS. A review of rice authenticity/adulteration methods and results. Crit Rev Food Sci Nutr. 2008;48:553–98.
Burgess K, Rankin N, Weidt S. Chapter 10 - Metabolomics. In: Padmanabhan S, editor. Handbook of pharmacogenomics and stratified medicine. San Diego, CA: Academic Press; 2014. p. 181–205. https://www.sciencedirect.com/science/article/pii/B9780123868824000104.
Lamichhane S, Sen P, Dickens AM, Hyötyläinen T, Orešič M. Chapter Fourteen - An overview of metabolomics data analysis: current tools and future perspectives. In: Jaumot J, Bedia C, Tauler R, editors. Comprehensive analytical chemistry. Amsterdam: Elsevier; 2018. p. 387–413. https://www.sciencedirect.com/science/article/pii/S0166526X18300655.
Fernie AR, Tadmor Y, Zamir D. Natural genetic variation for improving crop quality. Curr Opin Plant Biol. 2006;9:196–202.
Ashikari M, Matsuoka M. Identification, isolation and pyramiding of quantitative trait loci for rice breeding. Trends Plant Sci. 2006;11:344–50.
Yonekura-Sakakibara K, Saito K. Review: genetically modified plants for the promotion of human health. Biotechnol Lett. 2006;28:1983–91.
Hsing Y-I, Chern C-G, Fan M-J, Lu P-C, Chen K-T, Lo S-F, et al. A rice gene activation/knockout mutant resource for high throughput functional genomics. Plant Mol Biol. 2007;63:351–64.
Oikawa A, Matsuda F, Kusano M, Okazaki Y, Saito K. Rice metabolomics. Rice. 2008;1:63–71.
Ashikari M. Cytokinin oxidase regulates rice grain production. Science. 2005;309:741–5.
Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815.
Goff SA. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science. 2002;296:92–100.
Yu J. A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science. 2002;296:79–92.
Sasaki T, Matsumoto T, Yamamoto K, Sakata K, Baba T, Katayose Y, et al. The genome sequence and structure of rice chromosome 1. Nature. 2002;420:312–6.
International Rice Genome Sequencing Project. The map-based sequence of the rice genome. Nature. 2005;436:793–800.
Fiehn O. Metabolomics – the link between genotypes and phenotypes. Plant Mol Biol. 2002;48:155–71.
Dixon RA, Strack D. Phytochemistry meets genome analysis, and beyond. Phytochemistry. 2003;62:815–6.
Bino RJ, Hall RD, Fiehn O, Kopka J, Saito K, Draper J, et al. Potential of metabolomics as a functional genomics tool. Trends Plant Sci. 2004;9:418–25.
Fiehn O, Kopka J, Dörmann P, Altmann T, Trethewey RN, Willmitzer L. Metabolite profiling for plant functional genomics. Nat Biotechnol. 2000;18:1157–61.
Sumner LW, Mendes P, Dixon RA. Plant metabolomics: large-scale phytochemistry in the functional genomics era. Phytochemistry. 2003;62:817–36.
Fernie AR, Trethewey RN, Krotzky AJ, Willmitzer L. Metabolite profiling: from diagnostics to systems biology. Nat Rev Mol Cell Biol. 2004;5:763–9.
Fukusaki E, Kobayashi A. Plant metabolomics: potential for practical operation. J Biosci Bioeng. 2005;100:347–54.
Saito K, Dixon RA, Willmitzer L, editors. Plant metabolomics. Berlin: Springer; 2006. https://doi.org/10.1007/3-540-29782-0.
Fiehn O. Metabolite profiling in Arabidopsis. Methods Mol Biol. 2006;323:439–47.
Hollywood K, Brison DR, Goodacre R. Metabolomics: current technologies and future trends. Proteomics. 2006;6:4716–23.
Lisec J, et al. Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protoc. 2006;1:387–96.
Chen C, Gonzalez FJ, Idle JR. LC-MS-based metabolomics in drug metabolism. Drug Metab Rev. 2007;39:581–97.
Dettmer K, Aronov PA, Hammock BD. Mass spectrometry-based metabolomics. Mass Spectrom Rev. 2007;26:51–78.
Saito K, Hirai MY, Yonekura-Sakakibara K. Decoding genes with coexpression networks and metabolomics – ‘majority report by precogs’. Trends Plant Sci. 2008;13:36–43.
Aharoni A, Ric de Vos CH, Verhoeven HA, Maliepaard CA, Kruppa G, Bino R, et al. Nontargeted metabolome analysis by use of Fourier Transform Ion Cyclotron Mass Spectrometry. OMICS. 2002;6:217–34.
Soga T, Ohashi Y, Ueno Y, Naraoka H, Tomita M, Nishioka T. Quantitative metabolome analysis using capillary electrophoresis mass spectrometry. J Proteome Res. 2003;2:488–94.
Kristensen C, Morant M, Olsen CE, Ekstrøm CT, Galbraith DW, Møller BL, et al. Metabolic engineering of dhurrin in transgenic Arabidopsis plants with marginal inadvertent effects on the metabolome and transcriptome. Proc Natl Acad Sci U S A. 2005;102:1779–84.
Moco S, Bino RJ, Vorst O, Verhoeven HA, de Groot J, van Beek TA, et al. A Liquid chromatography-mass spectrometry-based metabolome database for tomato. Plant Physiol. 2006;141:1205–18.
Sato S, Soga T, Nishioka T, Tomita M. Simultaneous determination of the main metabolites in rice leaves using capillary electrophoresis mass spectrometry and capillary electrophoresis diode array detection. Plant J. 2004;40:151–63.
Takahashi H, Hayashi M, Goto F, Sato S, Soga T, Nishioka T, et al. Evaluation of metabolic alteration in transgenic rice overexpressing dihydroflavonol-4-reductase. Ann Bot. 2006;98:819–25.
Oikawa A, Nakamura Y, Ogura T, Kimura A, Suzuki H, Sakurai N, et al. Clarification of pathway-specific inhibition by fourier transform ion cyclotron resonance/mass spectrometry-based metabolic phenoty** studies. Plant Physiol. 2006;142:398–413.
Nakamura Y, Kimura A, Saga H, Oikawa A, Shinbo Y, Kai K, et al. Differential metabolomics unraveling light/dark regulation of metabolic activities in Arabidopsis cell culture. Planta. 2007;227:57–66.
Kusano M, Fukushima A, Kobayashi M, Hayashi N, Jonsson P, Moritz T, et al. Application of a metabolomic method combining one-dimensional and two-dimensional gas chromatography-time-of-flight/mass spectrometry to metabolic phenoty** of natural variants in rice. J Chromatogr B. 2007;855:71–9.
Krishnan P, Kruger NJ, Ratcliffe RG. Metabolite fingerprinting and profiling in plants using NMR. J Exp Bot. 2005;56:255–65.
Sekiyama Y, Kikuchi J. Towards dynamic metabolic network measurements by multi-dimensional NMR-based fluxomics. Phytochemistry. 2007;68:2320–9.
Overy SA, Walker HJ, Malone S, Howard TP, Baxter CJ, Sweetlove LJ, et al. Application of metabolite profiling to the identification of traits in a population of tomato introgression lines. J Exp Bot. 2005;56:287–96.
Dunn WB, Overy S, Quick WP. Evaluation of automated electrospray-TOF mass spectrometry for metabolic fingerprinting of the plant metabolome. Metabolomics. 2005;1:137–48.
Johnson HE, Broadhurst D, Kell DB, Theodorou MK, Merry RJ, Griffith GW. High-throughput metabolic fingerprinting of legume silage fermentations via fourier transform infrared spectroscopy and chemometrics. Appl Environ Microbiol. 2004;70:1583–92.
Suzuki H, Sasaki R, Ogata Y, Nakamura Y, Sakurai N, Kitajima M, et al. Metabolic profiling of flavonoids in Lotus japonicus using liquid chromatography Fourier transform ion cyclotron resonance mass spectrometry. Phytochemistry. 2008;69:99–111.
Iijima Y, Nakamura Y, Ogata Y, Tanaka K, Sakurai N, Suda K, et al. Metabolite annotations based on the integration of mass spectral information. Plant J. 2008;54:949–62.
Dwivedi P, Wu P, Klopsch SJ, Puzon GJ, Xun L, Hill HH. Metabolic profiling by ion mobility mass spectrometry (IMMS). Metabolomics. 2008;4:63–80.
Matsuda F, Okazaki Y, Oikawa A, Kusano M, Nakabayashi R, Kikuchi J, et al. Dissection of genotype–phenotype associations in rice grains using metabolome quantitative trait loci analysis. Plant J. 2012;70:624–36.
Routaboul J-M, Dubos C, Beck G, Marquis C, Bidzinski P, Loudet O, et al. Metabolite profiling and quantitative genetics of natural variation for flavonoids in Arabidopsis. J Exp Bot. 2012;63:3749–64.
Shi T, Zhu A, Jia J, Hu X, Chen J, Liu W, et al. Metabolomics analysis and metabolite-agronomic trait associations using kernels of wheat (Triticum aestivum) recombinant inbred lines. Plant J. 2020;103:279–92.
Wen W, Li K, Alseekh S, Omranian N, Zhao L, Zhou Y, et al. Genetic Determinants of the network of primary metabolism and their relationships to plant performance in a maize recombinant inbred line population. Plant Cell. 2015;27:1839–56.
Hill CB, Taylor JD, Edwards J, Mather D, Bacic A, Langridge P, et al. Whole-genome map** of agronomic and metabolic traits to identify novel quantitative trait loci in bread wheat grown in a water-limited environment. Plant Physiol. 2013;162:1266–81.
Li K, Wen W, Alseekh S, Yang X, Guo H, Li W, et al. Large-scale metabolite quantitative trait locus analysis provides new insights for high-quality maize improvement. Plant J. 2019;99:216–30.
Carreno-Quintero N, Acharjee A, Maliepaard C, Bachem CWB, Mumm R, Bouwmeester H, et al. Untargeted metabolic quantitative trait loci analyses reveal a relationship between primary metabolism and potato tuber quality. Plant Physiol. 2012;158:1306–18.
Nunes-Nesi A, Alseekh S, de Oliveira Silva FM, Omranian N, Lichtenstein G, Mirnezhad M, et al. Identification and characterization of metabolite quantitative trait loci in tomato leaves and comparison with those reported for fruits and seeds. Metabolomics. 2019;15:46. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6420416/.
Vallarino JG, Pott DM, Cruz-Rus E, Miranda L, Medina-Minguez JJ, Valpuesta V, et al. Identification of quantitative trait loci and candidate genes for primary metabolite content in strawberry fruit. Hortic Res. 2019;6:1–17.
Labadie M, Vallin G, Petit A, Ring L, Hoffmann T, Gaston A, et al. Metabolite quantitative trait loci for flavonoids provide new insights into the genetic architecture of strawberry (Fragaria × ananassa) fruit quality. J Agric Food Chem. 2020;68:6927–39.
Feng J, Long Y, Shi L, Shi J, Barker G, Meng J. Characterization of metabolite quantitative trait loci and metabolic networks that control glucosinolate concentration in the seeds and leaves of Brassica napus. New Phytol. 2012;193:96–108.
Piasecka A, Sawikowska A, Kuczyńska A, Ogrodowicz P, Mikołajczak K, Krystkowiak K, et al. Drought-related secondary metabolites of barley (Hordeum vulgare L.) leaves and their metabolomic quantitative trait loci. Plant J. 2017;89:898–913.
Alseekh S, Tohge T, Wendenberg R, Scossa F, Omranian N, Li J, et al. Identification and mode of inheritance of quantitative trait loci for secondary metabolite abundance in tomato. Plant Cell. 2015;27:485–512.
Hamzehzarghani H, Paranidharan V, Abu-Nada Y, Kushalappa AC, Mamer O, Somers D. Metabolic profiling to discriminate wheat near isogenic lines, with quantitative trait loci at chromosome 2DL, varying in resistance to fusarium head blight. Can J Plant Sci. 2011;88:789. https://doi.org/10.4141/CJPS07209.
Dong X, Gao Y, Chen W, Wang W, Gong L, Liu X, et al. Spatiotemporal distribution of phenolamides and the genetics of natural variation of hydroxycinnamoyl spermidine in rice. Mol Plant. 2015;8:111–21.
Chen W, Gao Y, **e W, Gong L, Lu K, Wang W, et al. Genome-wide association analyses provide genetic and biochemical insights into natural variation in rice metabolism. Nat Genet. 2014;46:714–21.
Riedelsheimer C, Lisec J, Czedik-Eysenberg A, Sulpice R, Flis A, Grieder C, et al. Genome-wide association map** of leaf metabolic profiles for dissecting complex traits in maize. Proc Natl Acad Sci U S A. 2012;109:8872–7.
Slaten ML, Yobi A, Bagaza C, Chan YO, Shrestha V, Holden S, et al. mGWAS uncovers Gln-glucosinolate seed-specific interaction and its role in metabolic homeostasis. Plant Physiol. 2020;183:483–500.
Joseph B, Corwin JA, Li B, Atwell S, Kliebenstein DJ. Cytoplasmic genetic variation and extensive cytonuclear interactions influence natural variation in the metabolome. elife. 2013;2:e00776.
Chen J, Hu X, Shi T, Yin H, Sun D, Hao Y, et al. Metabolite-based genome-wide association study enables dissection of the flavonoid decoration pathway of wheat kernels. Plant Biotechnol J. 2020;18:1722–35.
Ying S, Su M, Wu Y, Zhou L, Fu R, Li Y, et al. Trichome regulator SlMIXTA-like directly manipulates primary metabolism in tomato fruit. Plant Biotechnol J. 2020;18:354–63.
Angelovici R, Lipka AE, Deason N, Gonzalez-Jorge S, Lin H, Cepela J, et al. Genome-wide analysis of branched-chain amino acid levels in arabidopsis seeds. Plant Cell. 2013;25:4827–43.
Li H, Peng Z, Yang X, Wang W, Fu J, Wang J, et al. Genome-wide association study dissects the genetic architecture of oil biosynthesis in maize kernels. Nat Genet. 2013;45:43–50.
Shen M, Broeckling CD, Chu EY, Ziegler G, Baxter IR, Prenni JE, et al. Leveraging non-targeted metabolite profiling via statistical genomics. PLoS One. 2013;8:e57667.
Papageorgiou L, Eleni P, Raftopoulou S, Mantaiou M, Megalooikonomou V, Vlachakis D. Genomic big data hitting the storage bottleneck. EMBnet J. 2018;24:e910. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5958914/.
Arita M. Additional paper: computational resources for metabolomics. Brief Funct Genomic Proteomic. 2004;3:84–93.
Codrea MC, Jiménez CR, Heringa J, Marchiori E. Tools for computational processing of LC–MS datasets: a user’s perspective. Comput Methods Prog Biomed. 2007;86:281–90.
Shinbo Y, Nakamura Y, Altaf-Ul-Amin M, Asahi H, Kurokawa K, Arita M, et al. KNApSAcK: a comprehensive species-metabolite relationship database. In: Saito K, Dixon RA, Willmitzer L, editors. Plant metabolomics. Berlin: Springer; 2006. p. 165–81. https://doi.org/10.1007/3-540-29782-0_13.
Takahashi H, Hotta Y, Hayashi M, Kawai-Yamada M, Komatsu S, Uchimiya H. High throughput metabolome and proteome analysis of transgenic rice plants (Oryza sativa L.). Plant Biotechnol. 2005;22:47–50.
Catchpole GS, Beckmann M, Enot DP, Mondhe M, Zywicki B, Taylor J, et al. Hierarchical metabolomics demonstrates substantial compositional similarity between genetically modified and conventional potato crops. Proc Natl Acad Sci U S A. 2005;102:14458–62.
Baker JM, Hawkins ND, Ward JL, Lovegrove A, Napier JA, Shewry PR, et al. A metabolomic study of substantial equivalence of field-grown genetically modified wheat. Plant Biotechnol J. 2006;4:381–92.
Dixon RA, Gang DR, Charlton AJ, Fiehn O, Kuiper HA, Reynolds TL, et al. Applications of metabolomics in agriculture. Agric Food Chem. 2006;54:8984–94.
Kim JK, Bamba T, Harada K, Fukusaki E, Kobayashi A. Time-course metabolic profiling in Arabidopsis thaliana cell cultures after salt stress treatment. J Exp Bot. 2007;58:415–24.
Moco S, Capanoglu E, Tikunov Y, Bino RJ, Boyacioglu D, Hall RD, et al. Tissue specialization at the metabolite level is perceived during the development of tomato fruit. J Exp Bot. 2007;58:4131–46.
Parveen I, Moorby JM, Fraser MD, Allison GG, Kopka J. Application of gas chromatography–mass spectrometry metabolite profiling techniques to the analysis of heathland plant diets of sheep. J Agric Food Chem. 2007;55:1129–38.
Grata E, Boccard J, Glauser G, Carrupt P-A, Farmer EE, Wolfender J-L, et al. Development of a two-step screening ESI-TOF-MS method for rapid determination of significant stress-induced metabolome modifications in plant leaf extracts: the wound response in Arabidopsis thaliana as a case study. J Sep Sci. 2007;30:2268–78.
Jonsson P, Gullberg J, Nordström A, Kusano M, Kowalczyk M, Sjöström M, et al. A strategy for identifying differences in large series of metabolomic samples analyzed by GC/MS. Anal Chem. 2004;76:1738–45.
Hirai MY, Yano M, Goodenowe DB, Kanaya S, Kimura T, Awazuhara M, et al. Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2004;101:10205–10.
Hirai MY, Klein M, Fujikawa Y, Yano M, Goodenowe DB, Yamazaki Y, et al. Elucidation of gene-to-gene and metabolite-to-gene networks in arabidopsis by integration of metabolomics and transcriptomics. J Biol Chem. 2005;280:25590–5.
Kok EJ, Kuiper HA. Comparative safety assessment for biotech crops. Trends Biotechnol. 2003;21:439–44.
Kojima Y, Ebana K, Fukuoka S, Nagamine T, Kawase M. Development of an RFLP-based rice diversity research set of germplasm. Breed Sci. 2005;55:431–40.
Cevallos-Cevallos JM, Reyes-De-Corcuera JI, Etxeberria E, Danyluk MD, Rodrick GE. Metabolomic analysis in food science: a review. Trends Food Sci Technol. 2009;20:557–66.
Fernie AR, Schauer N. Metabolomics-assisted breeding: a viable option for crop improvement? Trends Genet. 2009;25:39–48.
Hu C, Shi J, Quan S, Cui B, Kleessen S, Nikoloski Z, et al. Metabolic variation between japonica and indica rice cultivars as revealed by non-targeted metabolomics. Sci Rep. 2014;4:5067.
Lawas LMF, Li X, Erban A, Kopka J, Jagadish SK, Zuther E, et al. Metabolic responses of rice cultivars with different tolerance to combined drought and heat stress under field conditions. GigaScience. 2019;8:giz050.
Tarpley L, Duran AL, Kebrom TH, Sumner LW. Biomarker metabolites capturing the metabolite variance present in a rice plant developmental period. BMC Plant Biol. 2005;5:1–12.
Fan TW-M, Lane AN, Shenker M, Bartley JP, Crowley D, Higashi RM. Comprehensive chemical profiling of gramineous plant root exudates using high-resolution NMR and MS. Phytochemistry. 2001;57:209–21.
Ghiasvand AR, Setkova L, Pawliszyn J. Determination of flavour profile in Iranian fragrant rice samples using cold-fibre SPME–GC–TOF–MS. Flav Frag J. 2007;22:377–91.
Kusano M, Yang Z, Okazaki Y, Nakabayashi R, Fukushima A, Saito K. Using metabolomic approaches to explore chemical diversity in rice. Mol Plant. 2015;8:58–67.
Zuther E, Koehl K, Kopka J. Comparative metabolome analysis of the salt response in breeding cultivars of rice. In: Advances in molecular breeding toward drought and salt tolerant crops. New York, NY: Springer; 2007. p. 285–315.
Chen W, Gong L, Guo Z, Wang W, Zhang H, Liu X, et al. A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: application in the study of rice metabolomics. Mol Plant. 2013;6:1769–80.
Schauer N, Fernie AR. Plant metabolomics: towards biological function and mechanism. Trends Plant Sci. 2006;11:508–16.
Li J, Niu X, Pei G, Sui X, Zhang X, Chen L, et al. Identification and metabolomic analysis of chemical modulators for lipid accumulation in Crypthecodinium cohnii. Bioresour Technol. 2015;191:362–8.
Allwood JW, Ellis DI, Goodacre R. Metabolomic technologies and their application to the study of plants and plant–host interactions. Physiol Plant. 2008;132:117–35.
Kueger S, Steinhauser D, Willmitzer L, Giavalisco P. High-resolution plant metabolomics: from mass spectral features to metabolites and from whole-cell analysis to subcellular metabolite distributions. Plant J. 2012;70:39–50.
Fiehn O. Combining genomics, metabolome analysis, and biochemical modelling to understand metabolic networks. Compar Funct Genom. 2001;2:155–68.
Nielsen J, Oliver S. The next wave in metabolome analysis. Trends Biotechnol. 2005;23:544–6.
Villas-Bôas SG, Mas S, Akesson M, Smedsgaard J, Nielsen J. Mass spectrometry in metabolome analysis. Mass Spectrom Rev. 2005;24:613–46.
Bolten CJ, Kiefer P, Letisse F, Portais J-C, Wittmann C. Sampling for metabolome analysis of microorganisms. Anal Chem. 2007;79:3843–9.
Razzaq A, Sadia B, Raza A, Khalid Hameed M, Saleem F. Metabolomics: a way forward for crop improvement. Metabolites. 2019;9:303.
Krasensky J, Jonak C. Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J Exp Bot. 2012;63:1593–608.
Fàbregas N, Fernie AR. The metabolic response to drought. J Exp Bot. 2019;70:1077–85.
Thomas V. Phenylpropanoid Biosynthesis. Mol Plant. 2010;3:2–20.
Todaka A, Umehara R, Sasaki K, Serizawa M, Urakami K, Kusuhara M, et al. Metabolic profiling of gemcitabine- and paclitaxel-treated immortalized human pancreatic cell lines with K-RASG12D. Biomed Res. 2017;38:29–40.
Nakabayashi R, Mori T, Saito K. Alternation of flavonoid accumulation under drought stress in Arabidopsis thaliana. Plant Signal Behav. 2014;9:e29518.
Wu H, Liu X, Zhang X, Ji C, Zhao J, Yu J. Proteomic and metabolomic responses of clam Ruditapes philippinarum to arsenic exposure under different salinities. Aquat Toxicol. 2013;136–137:91–100.
Gupta P, De B. Metabolomics analysis of rice responses to salinity stress revealed elevation of serotonin, and gentisic acid levels in leaves of tolerant varieties. Plant Signal Behav. 2017;12:e1335845.
Wanichthanarak K, Boonchai C, Kojonna T, Chadchawan S, Sangwongchai W, Thitisaksakul M. Deciphering rice metabolic flux reprograming under salinity stress via in silico metabolic modeling. Comput Struct Biotechnol J. 2020;18:3555–66.
Dhatt BK, Abshire N, Paul P, Hasanthika K, Sandhu J, Zhang Q, et al. Metabolic dynamics of develo** rice seeds under high night-time temperature stress. Front Plant Sci. 2019;10:1443. https://doi.org/10.3389/fpls.2019.01443.
Guy C, Kaplan F, Kopka J, Selbig J, Hincha DK. Metabolomics of temperature stress. Physiol Plant. 2008;132:220–35.
Yang M, Yang J, Su L, Sun K, Li D, Liu Y, et al. Metabolic profile analysis and identification of key metabolites during rice seed germination under low-temperature stress. Plant Sci. 2019;289:110282.
Vo KTX, Rahman MM, Rahman MM, Trinh KTT, Kim ST, Jeon J-S. Proteomics and metabolomics studies on the biotic stress responses of rice: an update. Rice. 2021;14:30.
Tugizimana F, Piater L, Dubery I. Plant metabolomics: a new frontier in phytochemical analysis. S Afr J Sci. 2013;109:01–11.
Hong J, Yang L, Zhang D, Shi J. Plant metabolomics: an indispensable system biology tool for plant science. Int J Mol Sci. 2016;17:767.
Niederbacher B, Winkler JB, Schnitzler JP. Volatile organic compounds as non-invasive markers for plant phenoty**. J Exp Bot. 2015;66:5403–16.
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Ethics declarations
None.
Rights and permissions
Copyright information
© 2021 The Author(s), under exclusive licence to Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Pati, P., Donde, R., Sabarinathan, S., Gouda, G., Gupta, M.K., Rathore, S.K. (2021). Metabolomics in Rice Improvement. In: Gupta, M.K., Behera, L. (eds) Applications of Bioinformatics in Rice Research. Springer, Singapore. https://doi.org/10.1007/978-981-16-3997-5_4
Download citation
DOI: https://doi.org/10.1007/978-981-16-3997-5_4
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-16-3996-8
Online ISBN: 978-981-16-3997-5
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)