Abstract
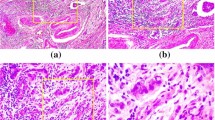
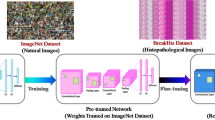
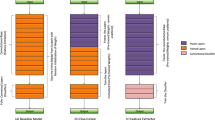
In the recent research, breast cancer has come out to be the biggest reason behind death among females. Detection of breast cancer in its earlier stages is really the need of time. Detection of cancerous tumor is really a long and time taking process which is very costly and requires a lot of workforce as well. Therefore, providing a computer assisted approach for the simpler classification areas could be a solution. This could simplify the complex examine process of pathologists. Hence, the most appropriate magnification factor based CNN framework is proposed which leads to lesser expenses and lesser workforce. The proposed framework considers the different magnification factor images to obtain the classification accuracy. The proposed CNN-based framework is sectioned into three parts: preprocessing, feature extraction based on CNN, and CNN-based classification. The preprocessing phase consists of four processes, i.e., resha**, formatting, image labeling, and train–test splitting. These preprocessings are applied after training and testing procedure of CNN model. In experimental analysis, publicly available breast cancer dataset from histopathology is used. The dataset consists of seven thousand nine hundred and nine images obtained from 82 different patients. These images are distributed among four different magnification factors: 40X, 100X, 200X, and 400X. These magnification factors are utilized for training of CNN model and for obtaining the accuracy. In result analysis, four different experimentations are conducted to show the effectualness of the presented CNN-based framework. Also, the results exhibit that the CNN-based framework used in this work achieves the highest average accuracy of 97.63% among the other architectures and existing approaches.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
T. Araújo, G. Aresta, E. Castro, J. Rouco, P. Aguiar, C. Eloy, A. Polónia, A. Campilho, Classification of breast cancer histology images using convolutional neural networks. PloS One 12(6), e0177544 (2017)
G. Aresta, T. Araújo, S. Kwok, S.S. Chennamsetty, M. Safwan, V. Alex, B. Marami, M. Prastawa, M. Chan, M. Donovan et al., BACH: grand challenge on breast cancer histology images. Med. Image Anal. 56, 122–139 (2019)
B.E. Bejnordi, M. Veta, P.J. Van Diest, B. Van Ginneken, N. Karssemeijer, G. Litjens, J.A. Van Der Laak, M. Hermsen, Q.F. Manson, M. Balkenhol et al., Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA 318(22), 2199–2210 (2017)
Y. Bengio, Practical recommendations for gradient-based training of deep architectures, Neural Networks: Tricks of the Trade (Springer, Berlin, 2012), pp. 437–478
Y.-L. Boureau, J. Ponce, Y. LeCun, A theoretical analysis of feature pooling in visual recognition, in Proceedings of the 27th International Conference on Machine Learning (ICML-10) (2010), pp. 111–118
H. Chen, Q. Dou, X. Wang, J. Qin, P.A. Heng, Mitosis detection in breast cancer histology images via deep cascaded networks, in 30th AAAI Conference on Artificial Intelligence (2016)
T. Ching, D.S. Himmelstein, B.K. Beaulieu-Jones, A.A. Kalinin, B.T. Do, G.P. Way, E. Ferrero, P.-M. Agapow, W. **e, G.L. Rosen et al., Opportunities and obstacles for deep learning in biology and medicine. biorxiv (2017)
D.C. Cireşan, A. Giusti, L.M. Gambardella, J. Schmidhuber, Mitosis detection in breast cancer histology images with deep neural networks, in International Conference on Medical Image Computing and Computer-Assisted Intervention (Springer, 2013), pp. 411–418
C. Désir, C. Petitjean, L. Heutte, M. Salaun, L. Thiberville, Classification of endomicroscopic images of the lung based on random subwindows and extra-trees. IEEE Trans. Biomed. Eng. 59(9), 2677–2683 (2012)
X. Glorot, Y. Bengio, Understanding the difficulty of training deep feedforward neural networks, in Proceedings of the 13th International Conference on Artificial Intelligence and Statistics (2010), pp. 249–256
M.N. Gurcan, L. Boucheron, A. Can, A. Madabhushi, N. Rajpoot, B. Yener, Histopathological image analysis: a review. IEEE Rev. Biomed. Eng. 2, 147 (2009)
N.A. Hamilton, R.S. Pantelic, K. Hanson, R.D. Teasdale, Fast automated cell phenotype image classification. BMC Bioinform. 8(1), 110 (2007)
V. Iglovikov, S. Mushinskiy, V. Osin, Satellite imagery feature detection using deep convolutional neural network: a Kaggle competition (2017), ar**v:1706.06169
V. Iglovikov, A. Shvets, TernausNet: U-Net with VGG11 encoder pre-trained on ImageNet for image segmentation (2018), ar**v:1801.05746
M. Kowal, P. Filipczuk, A. Obuchowicz, J. Korbicz, R. Monczak, Computer-aided diagnosis of breast cancer based on fine needle biopsy microscopic images. Comput. Biol. Med. 43(10), 1563–1572 (2013)
A. Krizhevsky, I. Sutskever, G.E. Hinton, ImageNet classification with deep convolutional neural networks, in Advances in Neural Information Processing Systems (2012), pp. 1097–1105
Y. Li, J. Wu, Q. Wu, Classification of breast cancer histology images using multi-size and discriminative patches based on deep learning. IEEE Access 7, 21400–21408 (2019)
G. Litjens, C.I. Sánchez, N. Timofeeva, M. Hermsen, I. Nagtegaal, I. Kovacs, C. Hulsbergen-Van De Kaa, P. Bult, B. Van Ginneken, J. Van Der Laak, Deep learning as a tool for increased accuracy and efficiency of histopathological diagnosis. Sci. Rep. 6, 26286 (2016)
A. Rakhlin, A. Shvets, V. Iglovikov, A.A. Kalinin, Deep convolutional neural networks for breast cancer histology image analysis, in International Conference Image Analysis and Recognition (Springer, 2018), pp. 737–744
S. Robertson, H. Azizpour, K. Smith, J. Hartman, Digital image analysis in breast pathology–from image processing techniques to artificial intelligence. Transl. Res. 194, 19–35 (2018)
J. Schmidhuber, Deep learning in neural networks: an overview. Neural Netw. 61, 85–117 (2015)
Y. Shin, I. Balasingham, Comparison of hand-craft feature based SVM and CNN based deep learning framework for automatic polyp classification, in 2017 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC) (IEEE, 2017), pp. 3277–3280
K. Simonyan, A. Zisserman, Very deep convolutional networks for large-scale image recognition (2014), ar**v:1409.1556
F.A. Spanhol, L.S. Oliveira, C. Petitjean, L. Heutte, A dataset for breast cancer histopathological image classification. IEEE Trans. Biomed. Eng. 63(7), 1455–1462 (2015)
F.A. Spanhol, L.S. Oliveira, C. Petitjean, L. Heutte, Breast cancer histopathological image classification using convolutional neural networks, in 2016 International Joint Conference on Neural Networks (IJCNN) (IEEE, 2016), pp. 2560–2567
C. Szegedy, W. Liu, Y. Jia, P. Sermanet, S. Reed, D. Anguelov, D. Erhan, V. Vanhoucke, A. Rabinovich, Going deeper with convolutions, in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (2015), pp. 1–9
D. Wang, A. Khosla, R. Gargeya, H. Irshad, A.H. Beck, Deep learning for identifying metastatic breast cancer (2016), ar**v:1606.05718
S.C. Wong, A. Gatt, V. Stamatescu, M.D. McDonnell, Understanding data augmentation for classification: when to warp? in 2016 International Conference on Digital Image Computing: Techniques and Applications (DICTA) (IEEE, 2016), pp. 1–6
Y. Yu, H. Lin, Q. Yu, J. Meng, Z. Zhao, Y. Li, L. Zuo, Modality classification for medical images using multiple deep convolutional neural networks. J. Comput. Inf. Syst. 11(15), 5403–5413 (2015)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Sharma, M., Verma, R., Mishra, A., Bhattacharya, M. (2020). A Novel Approach to Classify Breast Cancer Tumors Using Deep Learning Approach and Resulting Most Accurate Magnification Factor. In: Nanda, A., Chaurasia, N. (eds) High Performance Vision Intelligence. Studies in Computational Intelligence, vol 913. Springer, Singapore. https://doi.org/10.1007/978-981-15-6844-2_13
Download citation
DOI: https://doi.org/10.1007/978-981-15-6844-2_13
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-15-6843-5
Online ISBN: 978-981-15-6844-2
eBook Packages: EngineeringEngineering (R0)