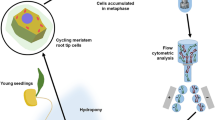
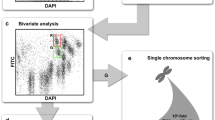
Abstract
Eukaryotic chromosomes can be studied either fixed on slides by use of cytogenetic techniques or through flow-cytometry. The latter enables not only the chromosome characterization but allows for the sorting and manipulation of individual chromosomes out of a complete genome, provided that target chromosomes have flow-cytometric distinctive features (e.g. size and/or DNA base content). These requirements represent a major constraint when the chromosomes of a given species are similarly sized, a common feature in plants. In wheat, this constraint can be overcome using special aneuploid mutants, each containing single arm pairs for a given chromosome of the complement, thus kee** a balanced genome composition while showing a complement with chromosome parts which are half-sized in respect to standard autosomes. However, such genotypes are available in hexaploid wheat (Triticum aestivum) exclusively in the background of Chinese Spring, a non-elite reference variety. Notably, aneuploids are not available for the most part of plants and animals. In order to overcome this major hurdle we have developed a reliable, fast and cost-effective method for Fluorescence labeling and In situ Hybridization of chromosomes In Suspension (FISHIS). The method makes use of fluorescent oligonucleotides of simple repetitive DNA sequences (SSR) or short DNA fragments, as probes, and allows for specific chromosome flow-sorting based on the hybridization pattern determined by the FISHIS probe. We have successfully applied the FISHIS methodology for flow karyoty** and sorting highly pure, single-type chromosome fractions of commercial varieties of bread and durum wheat and other Triticeae species. Moreover, the complete chromosomal sets corresponding to the two genomes (A and B) of durum wheat have also been clearly separated by the same FISHIS approach. Given the ubiquitous occurrence in eukaryotic genomes of the type of sequences used as probes, the abundance of their variants and their overall chromosome-specific distribution, their use in FISHIS experiments provides simple and fast access to individual chromosomes of virtually any eukaryotic genome, paving the way for gaining knowledge of great potential impact on basic and applied research aspects.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Ageno M, Dore E, Frontali C (1969) The alkaline denaturation of DNA. Biophys J 9:1281–1311
Arumuganathan K, Slattery JP, Tanksley SD, Earle ED (1991) Preparation and flow cytometric analysis of metaphase chromosomes of tomato. Theor Appl Gen 82:101–111
Bedbrook J, Jones JDG, O'Dell M, Thompson RD, Flavell RB (1980) Molecular description of telomeric heterochromatin in Secale species. Cell 19:545–560
Bento M, Gustafson JP, Viegas W, Silva M (2011) Size matters in Triticeae polyploids: larger genomes have higher remodeling. Genome 54:175–183
Bradbury L, Niehaus T, Hanson A (2013) Comparative genomics approaches to understanding and manipulating plant metabolism. Curr Opin Biotech 24:278–284
Brind’Amour J, Lansdorp PM (2011) Analysis of repetitive DNA in chromosomes by flow cytometry. Nat Methods 8:484–486
Carmona Á, Friero E, de Bustos A et al (2013) Cytogenetic diversity of SSR motifs within and between Hordeum species carrying the H genome: H. vulgare L. and H. bulbosum L. Theor Appl Genet 126:949–961
Chen J, Huang Q, Gao D et al (2013) Whole-genome sequencing of Oryza brachyantha reveals mechanisms underlying Oryza genome evolution. Nat Commun 4:1595
Chester M, Leitch AR, Soltis PS, Soltis DE (2010) Review of the application of modern cytogenetic methods (FISH/GISH) to the study of reticulation (Polyploidy/Hybridisation). Genes 1:166–192
Choy KW, Setlur SR, Lee C, Lau TK (2010) The impact of human copy number variation on a new era of genetic testing. BJOG 117:391–398
Claros MG, Bautista R, Guerrero-Fernández D et al (2012) Why assembling plant genome sequences is so challenging. Biology 1:439–459
Comai L (2005) The advantages and disadvantages of being polyploid. Nat Rev Genet 6:836–846
Conia J, Bergounioux C, Perennes C et al (1987) Flow cytometric analysis and sorting of plant chromosomes from Petunia hybrida protoplasts. Cytometry 8:500–508
Consortium TPGS (2011) Genome sequence and analysis of the tuber crop potato. Nature 475:189–195
Cook DR, Varshney RK (2010) From genome studies to agricultural biotechnology: closing the gap between basic plant science and applied agriculture. Curr Opin Plant Biol 13:115–118
Cuadrado Á, Schwarzacher T (1998) The chromosomal organization of simple sequence repeats in wheat and rye genomes. Chromosoma 107:587–594
Cuadrado Á, Schwarzacher T, Jouve N (2000) Identification of different chromatin classes in wheat using in situ hybridization with simple sequence repeat oligonucleotides. Theor Appl Genet 101:711–717
Cuadrado A, Jouve N (2007) The nonrandom distribution of long clusters of all possible classes of trinucleotide repeats in barley chromosomes. Chromosome Res 15:711–720
Cuadrado Á, Cardoso M, Jouve N (2008a) Increasing the physical markers of wheat chromosomes using SSRs as FISH probes. Genome 51:809–815
Cuadrado Á, Cardoso M, Jouve N (2008b) Physical organisation of simple sequence repeats (SSRs) in Triticeae: structural, functional and evolutionary implications. Cytogenet Genome Res 120:210–219
Cuadrado Á, Golczyk H, Jouve N (2009) A novel, simple and rapid nondenaturing FISH (ND-FISH) technique for the detection of plant telomeres. Potential used and possible target structures detected. Chromosome Res 17:755–762
Cuadrado A, Jouve N (2010) Chromosomal detection of simple sequence repeats (SSRs) using nondenaturing FISH (ND-FISH). Chromosoma 19:495–503
DeBolt S (2010) Copy number variation shapes genome diversity in Arabidopsis over immediate family generational scales. Genome Biol Evol 2:441–453
Delaat A, Blaas J (1984) Flow-cytometric characterization and sorting of plant chromosomes. Theor Appl Gen 67:463–467
Doležel J, Číhalíková J, Lucretti S (1992) A high-yield procedure for isolation of metaphase chromosomes from root-tips of Vicia faba L. Planta 188:93–98
Doležel J, Číhalíková J, Weiserova J, Lucretti S (1999) Cell cycle synchronization in plant root meristems. Methods Cell Sci 21:95–107
Doležel J, Kubaláková M, Paux E et al (2007) Chromosome-based genomics in the cereals. Chromosome Res 15:51–66
Doležel J, Šimková H, Kubaláková M et al (2009) Chromosome genomics in the Triticeae. In: Feuillet C, Muehlbauer GJ (eds) Genetics and genomics of the Triticeae. Springer ScienceþBusiness Media, Heidelberg, pp 385–316
Doležel J, Vrána J, Šafář J et al (2012) Chromosomes in the flow to simplify genome analysis. Funct Integr Genomic 12:397–416
Edwards D, Batley J, Snowdon RJ (2013) Accessing complex crop genomes with next-generation sequencing. Theor Appl Gen 126:1–11
Feuillet C, Leach JE, Rogers J et al (2011) Crop genome sequencing: lessons and rationales. Trends Plant Sci 16:77–88
Galbraith DW (2010) Flow cytometry and fluorescence-activated cell sorting in plants: the past, present, and future. Biomédica 30:65–70.
Galbraith DW, Lucretti S (2000) Large particle sorting. In: Radbruch A (ed) Flow cytometry and cell sorting. Springer Berlin Heidelberg, Heidelberg, pp 293–317
Gerlach W, Bedbrook J (1979) Sequence organization of the repeating units in the nucleus of wheat which contain 5s ribosomal-RNA genes. Nucleic Acids Res 8:4851–4865
Gill KS, Arumuganathan K, Lee JH (1999) Isolating individual wheat (Triticum aestivum) chromosome arms by flow cytometric analysis of ditelosomic lines. Theor App Gen 98:1248–1252
Giorgi D, Farina A, Grosso V et al (2013) FISHIS: Fluorescence In situ hybridization in suspension and chromosome flow sorting made easy. PLoS ONE 8:e57994
Greadzielewska A (2006) The genus Dasypyrum—part 2. Dasypyrum villosum—a wild species used in wheat improvement. Euphytica 152:441–454
Greilhuber J (2005) Intraspecific variation in genome size in angiosperms: identifying its existence. Ann Bot 95:91–98
Grosso V, Farina A, Gennaro A et al (2012) Flow sorting and molecular cytogenetic identification of individual chromosomes of Dasypyrum villosum L. (H. villosa) by a Single DNA Probe. PLoS ONE 7:e50151
Gualberti G, Doležel J, Macas J, Lucretti S (1996) Preparation of pea (Pisum sativum L) chromosome and nucleus suspensions from single root tips. Theor Appl Gen 92:744–751
Hansey CN, Vaillancourt B, Sekhon RS et al (2012) Maize (Zea mays L.) Genome Diversity as Revealed by RNA-Sequencing. PLoS ONE 7:e33071
He H, Deng W, Cassel MJ, Lucas JN (2001) Fluorescence in situ hybridization of metaphase chromosomes in suspension. Int J Radiat Biol 77:787–795
Henson J, Tischler G, Ning ZM (2012) Next-generation sequencing and large genome assemblies. Pharmacogenomics 13:901–915
Heslop-Harrison J, Schwarzacher T (2011) Organization of the plant genome in chromosomes. Plant J 66:18–33
Jiang J, Gill BS (2006) Current status and the future of fluorescence in situ hybridization (FISH) in plant genome research. Genome 49:1057–1068
Kalia RK, Rai MK, Kalia S et al (2011) Microsatellite markers: an overview of the recent progress in plants. Euphytica, 309–334
Kato A, Vega JM, Han F et al (2005) Advances in plant chromosome identification and cytogenetic techniques. Curr Opin Plant Biol 8:148–154
Kaul S, Koo HL, Jenkins J et al (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408:796–815
Kubaláková M, Vrána J, Číhalíková J et al (2002) Flow karyoty** and chromosome sorting in bread wheat (Triticum aestivum L.). Theor Appl Genet 104:1362–1372
Kubaláková M, Valarik M, Bartoš J et al (2003) Analysis and sorting of rye (Secale cereale L.) chromosomes using flow cytometry. Genome 46:893–905
Kubaláková M, Kovářová P, Suchánková P et al (2005) Chromosome Sorting in Tetraploid Wheat and Its Potential for Genome Analysis. Genetics 170:823–829
Lander ES, Linton LM, Birren B et al (2001) Initial sequencing and analysis of the human genome. Nature 409:860–921
Lee J-H, Ma Y, Wako T et al (2004) Flow karyotypes and chromosomal DNA contents of genus Triticum species and rye (Secale cereale). Chromosome Res 12:93–102
Li W, Chen P, Qi L, Liu D (1995) Isolation, characterization and application of a species-specific repeated sequence from Haynaldia villosa. Theor Appl Genet 90:526–533
Lucretti S, Doležel J (1995) Cell cycle synchronization, chromosome isolation, and flow-sorting in plants. Methods Cell Biol 50:61–83
Lucretti S, Doležel J, Schubert I, Fuchs J (1993) Flow karyoty** and sorting of Vicia faba chromosomes. Theor Appl Gen 85:665–672
Ma YZ, Lee JH, Li LC et al (2005) Fluorescent labeling of plant chromosomes in suspension by FISH. Genes Genet Syst 80:35–39
Macas J, Doležel J, Gualberti G et al (1995) Primer-induced labeling of pea and field bean chromosomes in situ and in suspension. Biotechniques 19:402–404; 407–408
Macas J, Doležel J, Lucretti S et al (1993) Localization of seed protein genes on flow-sorted field bean chromosomes. Chromosome Res 1:107–115
Mackay J, Dean JD, Plomion C et al (2012) Towards decoding the conifer giga-genome. Plant Mol Biol 80:555–569
Metzker ML (2010) Sequencing technologies: the next generation. Nat Rev Genet 11:31–46
Mizuno H, Wu J, Katayose Y et al (2008) Chromosome-specific distribution of nucleotide substitutions in telomeric repeats of rice (Oryza sativa L.). Mol Biol Evol 25:62–68
Molnár I, Kubaláková M, Simkova H et al (2011) Chromosome isolation by flow sorting in Aegilops umbellulata and Ae. comosa and their allotetraploid hybrids Ae. biuncialis and Ae. geniculata. PLoS One 6:e27708
Morgante M, De Paoli E, Radovic S (2007) Transposable elements and the plant pan-genomes. Curr Opin Plant Biol 10:149–155
Nie X, Li B, Wang L et al (2012) Development of chromosome-arm-specific microsatellite markers in Triticum aestivum (Poaceae) using NGS technology. Am J Bot 99:e369-e371
Page JT, Gingle AR, Udall JA (2013) PolyCat: a resource for genome categorization of sequencing reads from allopolyploid organisms. G3 3:517–525
Paterson AH, Bowers JE, Burow MD et al (2000) Comparative genomics of plant chromosomes. Plant Cell 12:1523–1539
Payseur BA, **g P, Haasl RJ (2011) A genomic portrait of human microsatellite variation. Mol Biol Evol 28:303–312
Pedersen C, Langridge P (1997) Identification of the entire chromosome complement of bread wheat by two-colour FISH. Genome 40:589–593
Pellicer J, Fay MF, Leitch IJ (2010) The largest eukaryotic genome of them all? Bot J Lin Soc 164:10–15
Pich U, Meister A, Macas J et al (1995) Primed in-situ labeling facilitates flow sorting of similar sized chromosomes. Plant J 7:1039–1044
Pruitt RE, Meyerowitz EM (1986) Characterization of the genome of Arabidopsis thaliana. J Mol Biol 187:169–183
Raap AK, Marijnen JG, Vrolijk J, Ploeg M van der (1986) Denaturation, renaturation, and loss of DNA during in situ hybridization procedures. Cytometry 7:235–242
Robertson KL, Thach DC (2009) LNA flow FISH: a flow cytometry fluorescence in situ hybridization method to detect messenger RNA using locked nucleic acid probes. Anal Biochem 390:109
Šafář J, Bartoš J, Janda J et al (2004) Dissecting large and complex genomes: flow sorting and BAC cloning of individual chromosomes from bread wheat. Plant J 39:960–968
Sanger F, Nicklen S, Coulson AR (1977) DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A 74:5463–5467
Schnable PS, Ware D, Fulton RS et al (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326:1112–1115
Sears E, Sears L (1978) The telocentric chromosomes of common wheat. In: Ramanujam S (ed) Indian Agricultural Research Institute, New Delhi, pp 389–407
Sharma S, Raina SN (2005) Organization and evolution of highly repeated satellite DNA sequence in plant chromosome. Cyt Gen Res 109:15–26
Šimková H, Svensson JT, Condamine P et al (2008) Coupling amplified DNA from flow-sorted chromosomes to high-density SNP map** in barley. BMC Genomics 9:294
Šmarda P, Bureš P, Horová L, Rotreklová O (2008) Intrapopulation genome size dynamics in Festuca pallens. Ann Botany 102:599–607
Smith DB, Flavell RB (1975) Characterisation of wheat genome by renaturation kinetics. Chromosoma 50:223–242
Speel EJM (1999) Detection and amplification systems for sensitive, multiple-target DNA and RNA in situ hybridization: Looking inside cells with a spectrum of colors. Hist Cell Biol 112:89–113
Steinhaeuser U, Starke H, Nietzel A et al (2002) Suspension (S)-FISH, a new technique for interphase nuclei. J Histochem Cytochem 50:1697–1698
Sutton T, Baumann U, Hayes J et al (2007) Boron-toxicity tolerance in barley arising from efflux transporter amplification. Science 318:1446–1449
Trask B, Vandenengh G, Landegent J et al (1985) Detection of dna-sequences in nuclei in suspension by in situ hybridization and dual beam flow-cytometry. Science 230:1401–1403
Treangen TJ, Salzberg SL (2012) Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat Genetics 13:36–46
Vandekken H, Arkesteijn GJA, Visser JWM, Bauman JGJ (1990) Flow cytometric quantification of human-chromosome specific repetitive dna-sequences by single and bicolor fluorescent insitu hybridization to lymphocyte interphase nuclei. Cytometry 11:153–164
Velasco R, Zharkikh A, Troggio M et al (2007) A high quality draft consensus sequence of the genome of a heterozygous grapevine variety. Plos One 2:e1326
Vitulo N, Albiero A, Forcato C et al (2011) First survey of the wheat chromosome 5A composition through a next generation sequencing approach. Plos One 6:e26421
Vrána J, Kubaláková M, Simkova H et al (2000) Flow sorting of mitotic chromosomes in common wheat (Triticum aestivum L.). Genetics 156:2033–2041
Wang XH, Lazzeri PA, Lorz H (1992) Chromosomal variation in dividing protoplasts derived from cell-suspensions of barley (Hordeum vulgare L). Theor Appl Gen 85:181–185
Wang Y, Wang X, Paterson AH (2012) Genome and gene duplications and gene expression divergence: a view from plants. Ann NY Acad Sci 1256:1–14
Wenzl P, Suchankova P, Carling J et al (2010) Isolated chromosomes as a new and efficient source of DArT markers for the saturation of genetic maps. Theor Appl Gen 121:465–474
Xu K, Xu X, Fukao T et al (2006) Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 442:705–708
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer Science+Business Media Dordrecht
About this chapter
Cite this chapter
Lucretti, S., Giorgi, D., Farina, A., Grosso, V. (2014). FISHIS: A New Way in Chromosome Flow Sorting Makes Complex Genomes More Accessible. In: Tuberosa, R., Graner, A., Frison, E. (eds) Genomics of Plant Genetic Resources. Springer, Dordrecht. https://doi.org/10.1007/978-94-007-7572-5_13
Download citation
DOI: https://doi.org/10.1007/978-94-007-7572-5_13
Published:
Publisher Name: Springer, Dordrecht
Print ISBN: 978-94-007-7571-8
Online ISBN: 978-94-007-7572-5
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)