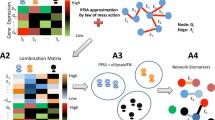
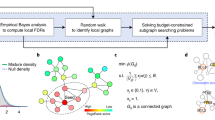
Abstract
Emerging research demonstrates the potential of protein-protein interaction (PPI) networks in uncovering the mechanistic bases of cancers, through identification of interacting proteins that are coordinately dysregulated in tumorigenic and metastatic samples. When used as features for classification, such coordinately dysregulated subnetworks improve diagnosis and prognosis of cancer considerably over single-gene markers. However, existing methods formulate coordination between multiple genes through additive representation of their expression profiles and utilize greedy heuristics to identify dysregulated subnetworks, which may not be well suited to the potentially combinatorial nature of coordinate dysregulation. Here, we propose a combinatorial formulation of coordinate dysregulation and decompose the resulting objective function to cast the problem as one of identifying subnetwork state functions that are indicative of phenotype. Based on this formulation, we show that coordinate dysregulation of larger subnetworks can be bounded using simple statistics on smaller subnetworks. We then use these bounds to devise an efficient algorithm, Crane, that can search the subnetwork space more effectively than simple greedy algorithms. Comprehensive cross-classification experiments show that subnetworks identified by Crane significantly outperform those identified by greedy algorithms in predicting metastasis of colorectal cancer (CRC).
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
Schadt, E.E., Lamb, J., Yang, X., Zhu, J., Edwards, S., Guhathakurta, D., Sieberts, S.K., Monks, S., Reitman, M., Zhang, C., Lum, P.Y., Leonardson, A., Thieringer, R., Metzger, J.M., Yang, L., Castle, J., Zhu, H., Kash, S.F., Drake, T.A., Sachs, A., Lusis, A.J.: An integrative genomics approach to infer causal associations between gene expression and disease. Nature Genetics 37(7), 710–717 (2005)
Papin, J.A., Hunter, T., Palsson, B.O., Subramaniam, S.: Reconstruction of cellular signalling networks and analysis of their properties. Nature Reviews Molecular Cell Biology 6(2), 99–111 (2005)
Ideker, T., Sharan, R.: Protein networks in disease. Genome Res. 18(4), 644–652 (2008)
Rich, J., Jones, B., Hans, C., Iversen, E., McClendon, R., Rasheed, A., Bigner, D., Dobra, A., Dressman, H., Nevins, J., West, M.: Gene expression profiling and genetic markers in glioblastoma survival. Cancer Research 65, 4051–4058 (2005)
Ewing, R.M., Chu, P., Elisma, F., Li, H., Taylor, P., Climie, S., McBroom-Cerajewski, L., Robinson, M.D., O’Connor, L., Li, M., Taylor, R., Dharsee, M., Ho, Y., Heilbut, A., Moore, L., Zhang, S., Ornatsky, O., Bukhman, Y.V., Ethier, M., Sheng, Y., Vasilescu, J., Abu-Farha, M., Lambert, J.P.P., Duewel, H.S., Stewart, I.I., Kuehl, B., Hogue, K., Colwill, K., Gladwish, K., Muskat, B., Kinach, R., Adams, S.L.L., Moran, M.F., Morin, G.B., Topaloglou, T., Figeys, D.: Large-scale map** of human protein-protein interactions by mass spectrometry. Molecular systems biology 3 (2007)
Goh, K.I., Cusick, M.E., Valle, D., Childs, B., Vidal, M., Barabasi, A.L.: The human disease network. PNAS 104(21), 8685–8690 (2007)
Rhodes, D.R., Chinnaiyan, A.M.: Integrative analysis of the cancer transcriptome. Nat. Genet. 37(suppl.) (June 2005)
Franke, L., Bakel, H., Fokkens, L., de Jong, E.D., Egmont-Petersen, M., Wijmenga, C.: Reconstruction of a functional human gene network, with an application for prioritizing positional candidate genes. Am. J. Hum. Genet. 78(6), 1011–1025 (2006)
Karni, S., Soreq, H., Sharan, R.: A network-based method for predicting disease-causing genes. Journal of Computational Biology 16(2), 181–189 (2009)
Lage, K., Karlberg, O.E., Størling, Z.M., Páll, P.A.G., Rigina, O., Hinsby, A.M., Tümer, Z., Pociot, F., Tommerup, N., Moreau, Y., Brunak, S.: A human phenome-interactome network of protein complexes implicated in genetic disorders. Nature Biotechnology 25(3), 309–316 (2007)
Ideker, T., Ozier, O., Schwikowski, B., Siegel, A.F.: Discovering regulatory and signalling circuits in molecular interaction networks. In: ISMB, pp. 233–240 (2002)
Guo, Z., Li, Y., Gong, X., Yao, C., Ma, W., Wang, D., Li, Y., Zhu, J., Zhang, M., Yang, D., Wang, J.: Edge-based scoring and searching method for identifying condition-responsive protein–protein interaction sub-network. Bioinformatics 23(16), 2121–2128 (2007)
Nacu, Ş., Critchley-Thorne, R., Lee, P., Holmes, S.: Gene expression network analysis and applications to immunology. Bioinformatics 23(7), 850–858 (2007)
Liu, M., Liberzon, A., Kong, S.W., Lai, W.R., Park, P.J., Kohane, I.S., Kasif, S.: Network-based analysis of affected biological processes in type 2 diabetes models. PLoS Genetics 3(6), e96 (2007)
Cabusora, L., Sutton, E., Fulmer, A., Forst, C.V.: Differential network expression during drug and stress response. Bioinformatics 21(12), 2898–2905 (2005)
Patil, K.R., Nielsen, J.: Uncovering transcriptional regulation of metabolism by using metabolic network topology. PNAS 102(8), 2685–2689 (2005)
Scott, M.S., Perkins, T., Bunnell, S., Pepin, F., Thomas, D.Y., Hallett, M.: Identifying regulatory subnetworks for a set of genes. Mol. Cell Prot., 683–692 (2005)
Chowdhury, S.A., Koyutürk, M.: Identification of coordinately dysregulated subnetworks in complex phenotypes. In: PSB, pp. 133–144 (2010)
Ulitsky, I., Karp, R.M., Shamir, R.: Detecting disease-specific dysregulated pathways via analysis of clinical expression profiles. In: Vingron, M., Wong, L. (eds.) RECOMB 2008. LNCS (LNBI), vol. 4955, pp. 347–359. Springer, Heidelberg (2008)
Chuang, H.Y., Lee, E., Liu, Y.T., Lee, D., Ideker, T.: Network-based classification of breast cancer metastasis. Mol. Syst. Biol. 3 (October 2007)
Nibbe, R.K., Ewing, R., Myeroff, L., Markowitz, M., Chance, M.: Discovery and scoring of protein interaction sub-networks discriminative of late stage human colon cancer. Mol. Cell Prot. 9(4), 827–845 (2009)
Nibbe, R.K., Koyutürk, M., Chance, M.R.: An integrative -omics approach to identify functional sub-networks in human colorectal cancer. PLoS Comput. Biol. 6(1), e1000639 (2010)
Anastassiou, D.: Computational analysis of the synergy among multiple interacting genes. Mol. Syst. Biol. 3(83) (2007)
Watkinson, J., Wang, X., Zheng, T., Anastassiou, D.: Identification of gene interactions associated with disease from gene expression data using synergy networks. BMC Systems Biology 2(1) (2008)
Quackenbush, J.: Microarray data normalization and transformation. Nat. Genet. 32(suppl.), 496–501 (2002)
Akutsu, T., Miyano, S., Kuhara, S.: Identification of genetic networks from a small number of gene expression patterns under the boolean network model. In: Pacific Symposium on Biocomputing, pp. 17–28 (1999)
Koyutürk, M., Szpankowski, W., Grama, A.: Biclustering gene-feature matrices for statistically significant dense patterns. In: IEEE Computational Systems Bioinformatics Conference (CSB 2004), pp. 480–484 (2004)
Akutsu, T., Miyano, S.: Selecting informative genes for cancer classification using gene expression data. In: Proceedings of the IEEE-EURASIP Workshop on Nonlinear Signal and Image Processing, pp. 3–6 (2001)
Shmulevich, I., Zhang, W.: Binary analysis and optimization-based normalization of gene expression data. Bioinformatics 18(4), 555–565 (2002)
Chowdhury, S.A., Nibbe, R.K., Chance, M.R., Koyutürk, M.: Supplement to “Subnetwork state functions define dysregulated subnetworks in cancer”, http://vorlon.case.edu/~mxk331/crane/recomb2010_supplement.pdf
Smyth, P., Goodman, R.M.: An information theoretic approach to rule induction from databases. IEEE Trans. on Knowl. and Data Eng. 4(4), 301–316 (1992)
Paschos, K., Canovas, D., Bird, N.: The role of cell adhesion molecules in the progression of colorectal cancer and the development of liver metastasis. Cell Signal 21(5), 665–674 (2009)
Zucker, S., Vacirca, J.: Role of matrix metalloproteinases (mmps) in colorectal cancer. Cancer Metastasis Rev. 23(1-2), 101–117 (2004)
McConnell, B., Yang, V.: The role of inflammation in the pathogenesis of colorectal cancer. Curr. Colorectal Cancer Rep. 5(2), 69–74 (2009)
Markowitz, S., Bertagnolli, M.: Molecular origins of cancer: Molecular basis of colorectal cancer. N. Engl. J. Med. 361(25), 2449–2460 (2009)
Vishnubhotla, R., Sun, S., Huq, J., Bulic, M., Ramesh, A.: Rock-ii mediates colon cancer invasion via regulation of mmp-2 and mmp-13 at the site of invadopodia as revealed by multiphoton imaging. Laboratory Investigation 87, 1149–1158 (2007)
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2010 Springer-Verlag Berlin Heidelberg
About this paper
Cite this paper
Chowdhury, S.A., Nibbe, R.K., Chance, M.R., Koyutürk, M. (2010). Subnetwork State Functions Define Dysregulated Subnetworks in Cancer. In: Berger, B. (eds) Research in Computational Molecular Biology. RECOMB 2010. Lecture Notes in Computer Science(), vol 6044. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-12683-3_6
Download citation
DOI: https://doi.org/10.1007/978-3-642-12683-3_6
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-12682-6
Online ISBN: 978-3-642-12683-3
eBook Packages: Computer ScienceComputer Science (R0)