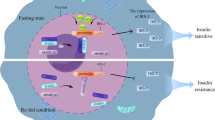
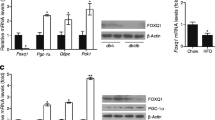
Abstract
As transcription factors, the forkhead box O family proteins control the expression of genes that are involved in the regulation of autophagy and metabolism. The FoxO–autophagy axis has been shown to mediate cell differentiation and tissue development. Dysregulated FoxO activity may compromise tissue development and homeostasis, concomitant with metabolic abnormalities across tissues such as liver, adipose tissue, skeletal muscle, and heart. In this chapter, we discuss the mechanism or pathways of FoxO transcription factors regulating autophagy and tissue integrity, and the FoxO–autophagy axis in cellular metabolism and fate determination. The evidence summarized here suggests that targeting the FoxO–autophagy axis may lead to therapeutic options for metabolic derangements and cell or tissue dysfunction.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
Abbreviations
- ACO:
-
Acyl-CoA oxidase
- Akt (or PKB):
-
Protein kinase B
- AMPK:
-
AMP-activated protein kinase
- Atg:
-
Autophagy related protein
- C/EBP:
-
CCAAT/enhancer-binding protein
- CD36:
-
Fatty acid translocase FAT/cluster of differentiation 36
- CMV:
-
Controlled mechanical ventilation
- CREB:
-
cAMP response element binding protein
- DBD:
-
DNA binding domain
- ER:
-
Endoplasmic reticulum
- FoxO:
-
Forkhead box O
- FSP27:
-
Fat-specific protein 27
- G6Pase:
-
Glucose-6-phosphatase
- HDAC:
-
Histone deacetylase
- HK:
-
Hexokinase
- JNK:
-
c-Jun N-terminal kinase
- KAA:
-
Ketogenic amino acid
- LC3:
-
Microtubule-associated protein 1A/1B-light chain 3-phosphatidylethanolamine conjugate
- LDHA:
-
Lactate dehydrogenase A
- LXR:
-
Liver X receptor
- MI:
-
Myocardial infarction
- MST1:
-
Mammalian sterile 20-like kinase 1
- mTORC1:
-
Mammalian target of rapamycin complex 1
- MTP:
-
Microsomal tryglyceride transfer protein
- MuRF1:
-
Muscle RING-finger protein-1
- NES:
-
Nuclear export sequence
- NLS:
-
Nuclear localization signal
- Pdx1:
-
Pancreas/duodenum homeobox gene-1
- PEPCK:
-
Phosphoenolpyruvate carboxykinase
- PGC1:
-
Peroxisome proliferator-activated receptor gamma coactivator 1
- PI3K:
-
Phosphatidylinositol 3 kinase
- PI3P:
-
Phoshpatidylinositol 3-phosphate
- PKA:
-
Protein kinase A
- PKM2:
-
Pyruvate kinase isozymes M2
- PPARγ:
-
Peroxisome proliferator-activated receptor γ
- RXR:
-
Retinoid X receptor
- Sirt1,2:
-
sirtuin 1, 2
- SKP2:
-
S-phase kinase-associated protein 2
- SQSTM1 (or p62):
-
Sequestosome 1
- SREBP:
-
Sterol response element-binding protein
- STZ:
-
Streptozotocin
- Tfeb:
-
Transcription factor EB
- ULK:
-
Unc-51-like kinase
- VLDL:
-
Very-low-density lipoprotein
- Vps34:
-
Vacuolar proteins 34
References
Jacobs FM, van der Heide LP, Wijchers PJ, Burbach JP, Hoekman MF, Smidt MP. FoxO6, a novel member of the FoxO class of transcription factors with distinct shuttling dynamics. J Biol Chem. 2003;278:35959–67.
Kaestner KH, Knochel W, Martinez DE. Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev. 2000;14:142–6.
Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–4.
Lin K, Dorman JB, Rodan A, Kenyon C. daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. 1997;278:1319–22.
Ogg S, Paradis S, Gottlieb S, et al. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–9.
Furuyama T, Nakazawa T, Nakano I, Mori N. Identification of the differential distribution patterns of mRNAs and consensus binding sequences for mouse DAF-16 homologues. Biochem J. 2000;349:629–34.
Obsil T, Obsilova V. Structure/function relationships underlying regulation of FOXO transcription factors. Oncogene. 2008;27:2263–75.
Cheng Z, White MF. Targeting Forkhead box O1 from the concept to metabolic diseases: lessons from mouse models. Antioxid Redox Signal. 2011;14:649–61.
van der Vos KE, Coffer PJ. The extending network of FOXO transcriptional target genes. Antioxid Redox Signal. 2011;14:579–92.
Matsuzaki H, Daitoku H, Hatta M, Tanaka K, Fukamizu A. Insulin-induced phosphorylation of FKHR (Foxo1) targets to proteasomal degradation. Proc Natl Acad Sci U S A. 2003;100:11285–90.
Huang H, Regan KM, Wang F, et al. Skp2 inhibits FOXO1 in tumor suppression through ubiquitin-mediated degradation. Proc Natl Acad Sci U S A. 2005;102:1649–54.
Brunet A, Sweeney LB, Sturgill JF, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–5.
Furuyama T, Kitayama K, Yamashita H, Mori N. Forkhead transcription factor FOXO1 (FKHR)-dependent induction of PDK4 gene expression in skeletal muscle during energy deprivation. Biochem J. 2003;375:365–71.
Nakae J, Kitamura T, Kitamura Y, Biggs WH 3rd, Arden KC, Accili D. The forkhead transcription factor Foxo1 regulates adipocyte differentiation. Dev Cell. 2003;4:119–29.
Zhang W, Patil S, Chauhan B, et al. FoxO1 regulates multiple metabolic pathways in the liver: effects on gluconeogenic, glycolytic, and lipogenic gene expression. J Biol Chem. 2006;281:10105–17.
Kim DH, Zhang T, Lee S, et al. FoxO6 integrates insulin signaling with MTP for regulating VLDL production in the liver. Endocrinology. 2014;155:1255–67.
Calabuig-Navarro V, Yamauchi J, Lee S, et al. Forkhead box O6 (FoxO6) depletion attenuates hepatic gluconeogenesis and protects against fat-induced glucose disorder in mice. J Biol Chem. 2015;290:15581–94.
Lee S, Dong HH. FoxO integration of insulin signaling with glucose and lipid metabolism. J Endocrinol. 2017;233:R67–79.
Zhou J, Liao W, Yang J, et al. FOXO3 induces FOXO1-dependent autophagy by activating the AKT1 signaling pathway. Autophagy. 2012;8:1712–23.
Zhao Y, Yang J, Liao W, et al. Cytosolic FoxO1 is essential for the induction of autophagy and tumour suppressor activity. Nat Cell Biol. 2010;12:665–75.
Zhao J, Brault JJ, Schild A, et al. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6:472–83.
Milan G, Romanello V, Pescatore F, et al. Regulation of autophagy and the ubiquitin-proteasome system by the FoxO transcriptional network during muscle atrophy. Nat Commun. 2015;6:6670.
Mammucari C, Milan G, Romanello V, et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6:458–71.
Chi Y, Shi C, Zhao Y, Guo C. Forkhead box O (FOXO) 3 modulates hypoxia-induced autophagy through AMPK signalling pathway in cardiomyocytes. Biosci Rep. 2016;36:e00345.
Liu L, Tao Z, Zheng LD, et al. FoxO1 interacts with transcription factor EB and differentially regulates mitochondrial uncoupling proteins via autophagy in adipocytes. Cell Death Dis. 2016;2:16066.
Liu L, Zheng LD, Zou P, et al. FoxO1 antagonist suppresses autophagy and lipid droplet growth in adipocytes. Cell Cycle. 2016;15:2033–41.
Lopategi A, Lopez-Vicario C, Alcaraz-Quiles J, et al. Role of bioactive lipid mediators in obese adipose tissue inflammation and endocrine dysfunction. Mol Cell Endocrinol. 2016;419:44–59.
Lanthier N, Leclercq IA. Adipose tissues as endocrine target organs. Best Pract Res Clin Gastroenterol. 2014;28:545–58.
Dowell P, Otto TC, Adi S, Lane MD. Convergence of peroxisome proliferator-activated receptor gamma and Foxo1 signaling pathways. J Biol Chem. 2003;278:45485–91.
Fan W, Imamura T, Sonoda N, et al. FOXO1 transrepresses peroxisome proliferator-activated receptor gamma transactivation, coordinating an insulin-induced feed-forward response in adipocytes. J Biol Chem. 2009;284:12188–97.
Armoni M, Harel C, Karni S, et al. FOXO1 represses peroxisome proliferator-activated receptor-gamma1 and -gamma2 gene promoters in primary adipocytes. A novel paradigm to increase insulin sensitivity. J Biol Chem. 2006;281:19881–91.
Nakae J, Cao Y, Oki M, et al. Forkhead transcription factor FoxO1 in adipose tissue regulates energy storage and expenditure. Diabetes. 2008;57:563–76.
**g E, Gesta S, Kahn CR. SIRT2 regulates adipocyte differentiation through FoxO1 acetylation/deacetylation. Cell Metab. 2007;6:105–14.
Bai L, Pang WJ, Yang YJ, Yang GS. Modulation of Sirt1 by resveratrol and nicotinamide alters proliferation and differentiation of pig preadipocytes. Mol Cell Biochem. 2008;307:129–40.
Higuchi M, Dusting GJ, Peshavariya H, et al. Differentiation of human adipose-derived stem cells into fat involves reactive oxygen species and Forkhead box O1 mediated upregulation of antioxidant enzymes. Stem Cells Dev. 2013;22:878–88.
Kim H, Hiraishi A, Tsuchiya K, Sakamoto K. (-) Epigallocatechin gallate suppresses the differentiation of 3T3-L1 preadipocytes through transcription factors FoxO1 and SREBP1c. Cytotechnology. 2010;62:245–55.
Munekata K, Sakamoto K. Forkhead transcription factor Foxo1 is essential for adipocyte differentiation. In Vitro Cell Dev Biol Anim. 2009;45:642–51.
Zou P, Liu L, Zheng L, et al. Targeting FoxO1 with AS1842856 suppresses adipogenesis. Cell Cycle. 2014;13:3759–67.
Gross DN, van den Heuvel AP, Birnbaum MJ. The role of FoxO in the regulation of metabolism. Oncogene. 2008;27:2320–36.
Samuel VT, Choi CS, Phillips TG, et al. Targeting foxo1 in mice using antisense oligonucleotide improves hepatic and peripheral insulin action. Diabetes. 2006;55:2042–50.
Titchenell PM, Chu Q, Monks BR, Birnbaum MJ. Hepatic insulin signalling is dispensable for suppression of glucose output by insulin in vivo. Nat Commun. 2015;6:7078.
Matsumoto M, Han S, Kitamura T, Accili D. Dual role of transcription factor FoxO1 in controlling hepatic insulin sensitivity and lipid metabolism. J Clin Invest. 2006;116:2464–72.
Kamagate A, Qu S, Perdomo G, et al. FoxO1 mediates insulin-dependent regulation of hepatic VLDL production in mice. J Clin Invest. 2008;118:2347–64.
Altomonte J, Cong L, Harbaran S, et al. Foxo1 mediates insulin action on apoC-III and triglyceride metabolism. J Clin Invest. 2004;114:1493–503.
Qu S, Su D, Altomonte J, et al. PPAR{alpha} mediates the hypolipidemic action of fibrates by antagonizing FoxO1. Am J Physiol Endocrinol Metab. 2007;292:E421–34.
Zhang K, Li L, Qi Y, et al. Hepatic suppression of Foxo1 and Foxo3 causes hypoglycemia and hyperlipidemia in mice. Endocrinology. 2012;153:631–46.
Tao R, **ong X, DePinho RA, Deng CX, Dong XC. Hepatic SREBP-2 and cholesterol biosynthesis are regulated by FoxO3 and Sirt6. J Lipid Res. 2013;54:2745–53.
de Lange P, Moreno M, Silvestri E, Lombardi A, Goglia F, Lanni A. Fuel economy in food-deprived skeletal muscle: signaling pathways and regulatory mechanisms. FASEB J. 2007;21:3431–41.
Sugita S, Kamei Y, Akaike F, et al. Increased systemic glucose tolerance with increased muscle glucose uptake in transgenic mice overexpressing RXRgamma in skeletal muscle. PLoS One. 2011;6:e20467.
Kamei Y, Mizukami J, Miura S, et al. A forkhead transcription factor FKHR up-regulates lipoprotein lipase expression in skeletal muscle. FEBS Lett. 2003;536:232–6.
Bastie CC, Nahle Z, McLoughlin T, et al. FoxO1 stimulates fatty acid uptake and oxidation in muscle cells through CD36-dependent and -independent mechanisms. J Biol Chem. 2005;280:14222–9.
Kamei Y, Miura S, Suganami T, et al. Regulation of SREBP1c gene expression in skeletal muscle: role of retinoid X receptor/liver X receptor and forkhead-O1 transcription factor. Endocrinology. 2008;149:2293–305.
Nakashima K, Yakabe Y. AMPK activation stimulates myofibrillar protein degradation and expression of atrophy-related ubiquitin ligases by increasing FOXO transcription factors in C2C12 myotubes. Biosci Biotechnol Biochem. 2007;71:1650–6.
Manfredi LH, Zanon NM, Garofalo MA, Navegantes LC. Kettelhut IC (2013) Effect of short-term cold exposure on skeletal muscle protein breakdown in rats. J Appl Physiol. 1985;115:1496–505.
Kamei Y, Miura S, Suzuki M, et al. Skeletal muscle FOXO1 (FKHR) transgenic mice have less skeletal muscle mass, down-regulated Type I (slow twitch/red muscle) fiber genes, and impaired glycemic control. J Biol Chem. 2004;279:41114–23.
Murtaugh LC. Pancreas and beta-cell development: from the actual to the possible. Development. 2007;134:427–38.
Straub SG, Sharp GW. Glucose-stimulated signaling pathways in biphasic insulin secretion. Diabetes Metab Res Rev. 2002;18:451–63.
Kitamura T, Nakae J, Kitamura Y, et al. The forkhead transcription factor Foxo1 links insulin signaling to Pdx1 regulation of pancreatic beta cell growth. J Clin Invest. 2002;110:1839–47.
Kobayashi M, Kikuchi O, Sasaki T, et al. FoxO1 as a double-edged sword in the pancreas: analysis of pancreas- and beta-cell-specific FoxO1 knockout mice. Am J Physiol Endocrinol Metab. 2012;302:E603–13.
Gupta D, Leahy AA, Monga N, Peshavaria M, Jetton TL, Leahy JL. Peroxisome proliferator-activated receptor gamma (PPARgamma) and its target genes are downstream effectors of FoxO1 protein in islet beta-cells: mechanism of beta-cell compensation and failure. J Biol Chem. 2013;288:25440–9.
Liu Y, Wang X, Wu H, et al. Glycine enhances muscle protein mass associated with maintaining Akt-mTOR-FOXO1 signaling and suppressing TLR4 and NOD2 signaling in piglets challenged with LPS. Am J Physiol Regul Integr Comp Physiol. 2016;311:R365–73.
Kitamura YI, Kitamura T, Kruse JP, et al. FoxO1 protects against pancreatic beta cell failure through NeuroD and MafA induction. Cell Metab. 2005;2:153–63.
Kawamori D, Kaneto H, Nakatani Y, et al. The forkhead transcription factor Foxo1 bridges the JNK pathway and the transcription factor PDX-1 through its intracellular translocation. J Biol Chem. 2006;281:1091–8.
Tanida I. Autophagosome formation and molecular mechanism of autophagy. Antioxid Redox Signal. 2011;14:2201–14.
Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–32.
Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–72.
Matsunaga K, Saitoh T, Tabata K, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11:385–96.
Obara K, Noda T, Niimi K, Ohsumi Y. Transport of phosphatidylinositol 3-phosphate into the vacuole via autophagic membranes in Saccharomyces cerevisiae. Genes Cells. 2008;13:537–47.
Sun Q, Fan W, Chen K, Ding X, Chen S, Zhong Q. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc Natl Acad Sci U S A. 2008;105:19211–6.
Sun Q, Fan W, Zhong Q. Regulation of Beclin 1 in autophagy. Autophagy. 2009;5:713–6.
Itakura E, Mizushima N. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy. 2010;6:764–76.
Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–41.
Egan DF, Shackelford DB, Mihaylova MM, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–61.
Mizushima N, Yamamoto A, Hatano M, et al. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152:657–68.
Hanada T, Noda NN, Satomi Y, et al. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem. 2007;282:37298–302.
Bjorkoy G, Lamark T, Brech A, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–14.
Pankiv S, Clausen TH, Lamark T, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–45.
Rogov V, Dotsch V, Johansen T, Kirkin V. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol Cell. 2014;53:167–78.
**ong X, Tao R, DePinho RA, Dong XC. The autophagy-related gene 14 (Atg14) is regulated by forkhead box O transcription factors and circadian rhythms and plays a critical role in hepatic autophagy and lipid metabolism. J Biol Chem. 2012;287:39107–14.
Sengupta A, Molkentin JD, Yutzey KE. FoxO transcription factors promote autophagy in cardiomyocytes. J Biol Chem. 2009;284:28319–31.
Settembre C, Di Malta C, Polito VA, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–33.
Settembre C, De Cegli R, Mansueto G, et al. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat Cell Biol. 2013;15:647–58.
Singh R, **ang Y, Wang Y, et al. Autophagy regulates adipose mass and differentiation in mice. J Clin Invest. 2009;119:3329–39.
Zhang Y, Goldman S, Baerga R, Zhao Y, Komatsu M, ** S. Adipose-specific deletion of autophagy-related gene 7 (atg7) in mice reveals a role in adipogenesis. Proc Natl Acad Sci U S A. 2009;106:19860–5.
Zhang C, He Y, Okutsu M, et al. Autophagy is involved in adipogenic differentiation by repressesing proteasome-dependent PPARgamma2 degradation. Am J Physiol Endocrinol Metab. 2013;305:E530–9.
Puri V, Ranjit S, Konda S, et al. Cidea is associated with lipid droplets and insulin sensitivity in humans. Proc Natl Acad Sci U S A. 2008;105:7833–8.
Wang H, Liu L, Lin JZ, Aprahamian TR, Farmer SR. Browning of white adipose tissue with roscovitine induces a distinct population of UCP1+ adipocytes. Cell Metab. 2016;24:835–47.
Taylor D, Gottlieb RA. Parkin-mediated mitophagy is downregulated in browning of white adipose tissue. Obesity (Silver Spring). 2017;25:704–12.
Kalinovich AV, de Jong JM, Cannon B, Nedergaard J. UCP1 in adipose tissues: two steps to full browning. Biochimie. 2017;134:127–37.
Xu L, Kanasaki M, He J, et al. Ketogenic essential amino acids replacement diet ameliorated hepatosteatosis with altering autophagy-associated molecules. Biochim Biophys Acta. 2013;1832:1605–12.
Song YM, Lee YH, Kim JW, et al. Metformin alleviates hepatosteatosis by restoring SIRT1-mediated autophagy induction via an AMP-activated protein kinase-independent pathway. Autophagy. 2015;11:46–59.
Ratti F, Ramond F, Moncollin V, et al. Histone deacetylase 6 is a FoxO transcription factor-dependent effector in skeletal muscle atrophy. J Biol Chem. 2015;290:4215–24.
O’Neill BT, Lee KY, Klaus K, et al. Insulin and IGF-1 receptors regulate FoxO-mediated signaling in muscle proteostasis. J Clin Invest. 2016;126:3433–46.
Brault JJ, Jespersen JG, Goldberg AL. Peroxisome proliferator-activated receptor gamma coactivator 1alpha or 1beta overexpression inhibits muscle protein degradation, induction of ubiquitin ligases, and disuse atrophy. J Biol Chem. 2010;285:19460–71.
Wei B, Dui W, Liu D, **ng Y, Yuan Z, Ji G. MST1, a key player, in enhancing fast skeletal muscle atrophy. BMC Biol. 2013;11:12.
Lee D, Goldberg AL. SIRT1 protein, by blocking the activities of transcription factors FoxO1 and FoxO3, inhibits muscle atrophy and promotes muscle growth. J Biol Chem. 2013;288:30515–26.
Bertaggia E, Coletto L, Sandri M. Posttranslational modifications control FoxO3 activity during denervation. Am J Physiol Cell Physiol. 2012;302:C587–96.
Hussain SN, Mofarrahi M, Sigala I, et al. Mechanical ventilation-induced diaphragm disuse in humans triggers autophagy. Am J Respir Crit Care Med. 2010;182:1377–86.
Machado J, Manfredi LH, Silveira WA, et al. Calcitonin gene-related peptide inhibits autophagic-lysosomal proteolysis through cAMP/PKA signaling in rat skeletal muscles. Int J Biochem Cell Biol. 2016;72:40–50.
Hariharan N, Maejima Y, Nakae J, Paik J, Depinho RA, Sadoshima J. Deacetylation of FoxO by Sirt1 plays an essential role in mediating starvation-induced autophagy in cardiac myocytes. Circ Res. 2010;107:1470–82.
Paula-Gomes S, Goncalves DA, Baviera AM, Zanon NM, Navegantes LC, Kettelhut IC. Insulin suppresses atrophy- and autophagy-related genes in heart tissue and cardiomyocytes through AKT/FOXO signaling. Horm Metab Res. 2013;45:849–55.
Zaglia T, Milan G, Franzoso M, et al. Cardiac sympathetic neurons provide trophic signal to the heart via beta2-adrenoceptor-dependent regulation of proteolysis. Cardiovasc Res. 2013;97:240–50.
Sengupta A, Molkentin JD, Paik JH, DePinho RA, Yutzey KE. FoxO transcription factors promote cardiomyocyte survival upon induction of oxidative stress. J Biol Chem. 2011;286:7468–78.
Ning Y, Li Z, Qiu Z. FOXO1 silence aggravates oxidative stress-promoted apoptosis in cardiomyocytes by reducing autophagy. J Toxicol Sci. 2015;40:637–45.
Siegrist SE, Haque NS, Chen CH, Hay BA, Hariharan IK. Inactivation of both Foxo and reaper promotes long-term adult neurogenesis in Drosophila. Curr Biol. 2010;20:643–8.
Xu P, Das M, Reilly J, Davis RJ. JNK regulates FoxO-dependent autophagy in neurons. Genes Dev. 2011;25:310–22.
Jia K, Thomas C, Akbar M, et al. Autophagy genes protect against Salmonella typhimurium infection and mediate insulin signaling-regulated pathogen resistance. Proc Natl Acad Sci U S A. 2009;106:14564–9.
Wang S, **a P, Huang G, et al. FoxO1-mediated autophagy is required for NK cell development and innate immunity. Nat Commun. 2016;7:11023.
Deng Y, Kerdiles Y, Chu J, et al. Transcription factor Foxo1 is a negative regulator of natural killer cell maturation and function. Immunity. 2015;42:457–70.
Demontis F, Perrimon N. FOXO/4E-BP signaling in Drosophila muscles regulates organism-wide proteostasis during aging. Cell. 2010;143:813–25.
Bai H, Kang P, Hernandez AM, Tatar M. Activin signaling targeted by insulin/dFOXO regulates aging and muscle proteostasis in Drosophila. PLoS Genet. 2013;9:e1003941.
Akasaki Y, Hasegawa A, Saito M, Asahara H, Iwamoto Y, Lotz MK. Dysregulated FOXO transcription factors in articular cartilage in aging and osteoarthritis. Osteoarthritis Cartilage. 2014;22:162–70.
Chiacchiera F, Matrone A, Ferrari E, et al. p38alpha blockade inhibits colorectal cancer growth in vivo by inducing a switch from HIF1alpha- to FoxO-dependent transcription. Cell Death Differ. 2009;16:1203–14.
Matrone A, Grossi V, Chiacchiera F, et al. p38alpha is required for ovarian cancer cell metabolism and survival. Int J Gynecol Cancer. 2010;20:203–11.
Zhang J, Ng S, Wang J, et al. Histone deacetylase inhibitors induce autophagy through FOXO1-dependent pathways. Autophagy. 2015;11:629–42.
Zhu WL, Tong H, Teh JT, Wang M. Forkhead box protein O3 transcription factor negatively regulates autophagy in human cancer cells by inhibiting forkhead box protein O1 expression and cytosolic accumulation. PLoS One. 2014;9:e115087.
Acknowledgments
This work was supported by USDA National Institute of Food and Agriculture Hatch Project 1007334 (ZC) and American Heart Association Grant 18TPA34230082 (ZC) . The authors declare no potential conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Liu, L., Cheng, Z. (2018). Forkhead Box O (FoxO) Transcription Factors in Autophagy, Metabolic Health, and Tissue Homeostasis. In: Turksen, K. (eds) Autophagy in Health and Disease. Stem Cell Biology and Regenerative Medicine. Humana Press, Cham. https://doi.org/10.1007/978-3-319-98146-8_4
Download citation
DOI: https://doi.org/10.1007/978-3-319-98146-8_4
Published:
Publisher Name: Humana Press, Cham
Print ISBN: 978-3-319-98145-1
Online ISBN: 978-3-319-98146-8
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)