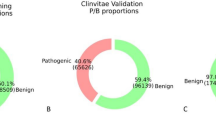
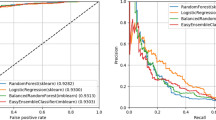
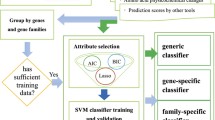
Abstract
The exome or genome based high throughput screening techniques are becoming a definitive criterion in the conventional clinical analysis of the genetic diseases. However, pathogenic classification of an identified variant, is still a manual and time consuming process for clinical geneticists. Thus, to facilitate the variant classification process, we have developed GeVaCT, a Java based tool that implements a classification approach based on the literature review of cardiac arrhythmia syndromes. Furthermore, the adoption of this automated knowledge engineer by the clinical geneticists will aid to build a knowledge base for the evolution of the variant classification process by use of novel machine learning approaches.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Saunders, C.J., et al.: Rapid whole-genome sequencing for genetic disease diagnosis in neonatal intensive care units. Sci. Transl. Med. 4, 135–154 (2012)
Hofman, N., et al.: Yield of molecular and clinical testing for arrhythmia syndromes: report of a 15 years’ experience. Circulation 1513–1521 (2013)
Schulze-Bahr, E., et al.: Molecular genetics of arrhythmias – a new paradigm. Z Kardiol. 89(4), IV12–IV22 (2000)
Wilde, A.A.M., Tan, H.L.: Inherited arrhythmia syndromes. Circ. J. 71(Supplement A), A12–A19 (2007)
Stenson, P.D., et al.: Human gene mutation database HGMD: 2003 update. Hum. Mutat. 21(6), 577–581 (2003)
Landrum, M.J., et al.: ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 42(D1), D980–D985 (2014)
Sherry, S.T., et al.: dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 29(1), 308–311 (2001)
Adzhubei, I.A., et al.: A method and server for predicting damaging missense mutations. Nat. Methods 7(4), 248–249 (2010)
Kumar, P., Henikoff, S., Ng, P.C.: Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 4(7), 1073–1081 (2009)
Mathe, E., et al.: Computational approaches for predicting the biological effect of p53 missense mutations: a comparison of three sequence analysis based methods. Nucleic Acids Res. 34(5), 1317–1325 (2006)
Pollard, K.S., Hubisz, M.J., Rosenbloom, K.R., Siepel, A.: Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 20(1), 110–121 (2010)
Exome Aggregation Consortium (2015). http://exac.broadinstitute.org
Desmet, F.-O., et al.: Human splicing finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 37(9), e67 (2009)
Plon, S.E., et al.: Sequence variant classification and reporting: recommendations for improving the interpretation of cancer susceptibility genetic test results. Hum. Mutat. 29(11), 1282–1291 (2008)
Waterman, D.A., Hayes-Roth, F.: An Investigation of Tools for Building Expert Systems, vol. 116. Addison-Wesley, Reading (1983)
Witten, I.H., Frank, E., Hall, M.A.: Data Mining Practical Machine Learning Tools and Techniques, 3rd edn. Morgan Kaufmann, Los Altos (2011)
Grau, I., et al.: GeVaCT: Genomic Variant Classifier Tool. http://bridgeiris.ulb.ac.be:81/gevact/
Acknowledgments
The authors acknowledge the support of the BridgeIRIS project funded by INNOVIRIS, Brussels, Belgium and the Cuba-Flanders VLIR Network.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer International Publishing AG
About this paper
Cite this paper
Grau, I. et al. (2018). Genomic Variant Classifier Tool. In: Bi, Y., Kapoor, S., Bhatia, R. (eds) Proceedings of SAI Intelligent Systems Conference (IntelliSys) 2016. IntelliSys 2016. Lecture Notes in Networks and Systems, vol 15. Springer, Cham. https://doi.org/10.1007/978-3-319-56994-9_32
Download citation
DOI: https://doi.org/10.1007/978-3-319-56994-9_32
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-56993-2
Online ISBN: 978-3-319-56994-9
eBook Packages: EngineeringEngineering (R0)