Abstract
We propose a new machine learning formulation designed specifically for extrapolation. The textbook way to apply machine learning to drug design is to learn a univariate function that when a drug (structure) is input, the function outputs a real number (the activity): F(drug) → activity. The PubMed server lists around twenty thousand papers doing this. However, experience in real-world drug design suggests that this formulation of the drug design problem is not quite correct. Specifically, what one is really interested in is extrapolation: predicting the activity of new drugs with higher activity than any existing ones. Our new formulation for extrapolation is based around learning a bivariate function that predicts the difference in activities of two drugs: F(drug1, drug2) → signed difference in activity. This formulation is general and potentially suitable for problems to find samples with target values beyond the target value range of the training set. We applied the formulation to work with support vector machines (SVMs), random forests (RFs), and Gradient Boosting Machines (XGBs). We compared the formulation with standard regression on thousands of drug design datasets, and hundreds of gene expression datasets. The test set extrapolation metrics use the concept of classification metrics to count the identification of extraordinary examples (with greater values than the training set), and top-performing examples (within the top 10% of the whole dataset). On these metrics our pairwise formulation vastly outperformed standard regression for SVMs, RFs, and XGBs. We expect this success to extrapolate to other extrapolation problems.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
The original motivation for this work came from applying machine learning (ML) to drug design, specifically quantitative structure activity relationship (QSAR) learning. The standard way to cast QSAR learning as ML is to learn a univariate function that when a drug (structure) is input, the function outputs a real number (the activity): F(drug) → activity. The PubMed server lists around twenty thousand papers doing this.
Experience in real-world drug discovery suggests that this formulation is not exactly what is really required in practice. Specifically, what one is really interested in is predicting the activity of new drugs with higher activity than any existing ones - extrapolation. N.B. extrapolation in QSAR learning has two related meanings: one is the ability to make predictions for molecules with descriptor values (xi) outside the applicability domain defined by the training set of the model (Fig. 1a) [1,2,3]; the other is the identification of the “extraordinary molecules” with activities (y) beyond the range of activity values in the training data (Fig. 1b) [1, 4]. In drug discovery both types of extrapolation are important. Extrapolating beyond training set descriptor values enables new molecular types (maybe unpatented) to be proposed. Extrapolating beyond the highest observed y values is strongly desired to select more effective drugs.
Although many QSAR learning studies have reported advantageous ML methods based on their model prediction accuracy using metrics such as mean squared error, in practice the ability to produce accurate predictions is less valuable than the extrapolation ability in this type of application [4, 5]. In fact, some ML methods can hardly extrapolate beyond the training sets. For example, random forest (RF) is incapable of predicting target values (y) outside the range of the training set because it gives ensembled prediction by averaging over its leaf predictions [4, 1]. Von Korff and Sander used sorted and shuffled datasets to evaluate extrapolation and interpolation performance, respectively [4]. **ong et al., Meredig et al. and Watson et al. have each proposed a new model validation technique to evaluate extrapolation performance of ML methods [6, 12, 13]. However, due to a lack of systematic review for them, it is unclear that these methods are statistically meaningful. Therefore, in this study, we apply standard k-fold cross validation [2, 6, 14].
This study proposes a ML configuration approach, the “pairwise approach”, to boost the extrapolation ability of a traditional regression learning method. A pairwise model is designed to model the relationship between the differences in the structures of pairs of drugs and the sign of differences in their activity values. The learned pairwise model is a bivariate function, F(drug1, drug2) → signed difference in activity, whose outputs can give a better ranking of drugs by ranking algorithms. By transforming the learning objective, the pairwise model enables improved performance in extrapolation compared to traditional regression evaluation.
2 Method
2.1 Datasets and Data Pre-processing
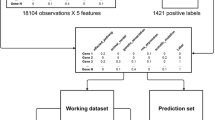
ChEMBL is a chemical database of bioactive molecules [15, 16]. It contains a large number of molecules and their measured activities against a variety of targets. Due to their size and scope, these datasets are suitable for benchmarking ML applications in the realm of QSAR [17]. ChEMBL features a number of different activities, in this study we are employing pXC50 as our target values, i.e. -log(measured activity). The structure of drug molecules is represented by the commonly employed Morgan fingerprint (1024 bits, r = 2) encoding the molecular substructures by Boolean values [18].
The other large-scale database we used is the human gene expression datasets (accession code GSE70138) from the Library of Integrated Network-based Cellular Signatures data (LINCS) [19]. These datasets were used by Olier et al. in transformational ML study [16]. This set of datasets contains the measured gene expression level across different tissue types and drug treatments in cancer cell lines. There are in total of 978 human genes, each of which was measured under 118,050 experimental conditions. Each dataset is the expression levels of a gene, measured and processed as level 5 differential gene expression signatures, under a series of conditions. The conditions are featured into 1,154 Boolean values describing drugs’ fingerprints (1024 bits) and experimental settings, which include 83 dosages, 14 cell types and 3 time points.
Before training any ML model, a basic feature selection is performed to reduce the large feature space and accelerate the learning process. For a given dataset, the features were removed if they have the same feature value assigned to every sample in a dataset. The features that repeat to have the same pattern for all the samples were also removed.
2.2 Formulation of Baseline Approach
In this study, when evaluating the performance of the pairwise approach on a specific dataset, in most cases it is compared with that of a baseline ML configuration, addressed as the “standard approach”. It refers to the standard way of learning a regression problem. For a dataset, the model is built directly between the feature vector, \({\mathbf{x}}_{{\varvec{i}}}\in {R}^{\left({N}_{f}\times {N}_{s}\right)}\), and the target value, \({y}_{{\varvec{i}}}\in {R}^{{N}_{s}}\) of all the samples. With multiple samples of known \(\left(\mathbf{x},y\right)\), a ML method can learn the relationship between features, \(\mathbf{x}\), and target values, \(y\), establishing a model \(f\) which can produce \(f({\mathbf{x}}_{{\varvec{i}}}) \approx\) \({y}_{i}\) for the training set. The feature values of the test samples, \({\mathbf{x}}_{ts}\) are fed into the model \(f\) to obtain the predicted target values, \({y}_{ts}^{\mathrm{pred}}=f({\mathbf{x}}_{ts})\). The performance of this model is then evaluated using metrics for evaluating the extrapolation performance (see Sect. 2.6).
2.3 Formulation of Pairwise Approach
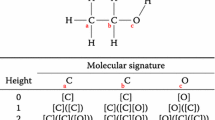
For a given Boolean dataset, a pair of samples \({P}_{\mathrm{AB}}\) is derived from sample A (\({S}_{\mathrm{A}}\)) and sample B (\({S}_{\mathrm{B}}\)). The difference in the ith feature for this pair can be presented in one of the following ways: present in both samples (\({x}_{\mathrm{A},i}\) = 1, \({x}_{\mathrm{B},i}\)= 1), present in \({S}_{\mathrm{A}}\) but not in \({S}_{\mathrm{B}}\) (\({x}_{\mathrm{A},i}\)= 1, \({x}_{\mathrm{B},i}\)= 0), present in \({S}_{\mathrm{B}}\) but not in \({S}_{\mathrm{A}}\) (\({x}_{\mathrm{A},i}\)= 0, \({x}_{\mathrm{B},i}\)= 1), and absent from both samples (\({x}_{\mathrm{A},i}\)= 0, \({x}_{\mathrm{B},i}\)= 0). To represent each type of difference in a feature, a unique value is assigned to the ith feature of the pair. An example of generating pairwise feature for the ith feature from a ChEMBL dataset is shown in Fig. 2. The unique values used in our experiments are:
The way of generating pairwise features is called ordinal encoding. It is often used for categorical features and each category value is assigned an integer value. Another popular way to encode real values for categorical features is one-hot encoding. It assigns Boolean bits to describe the absence or presence of each category. Therefore, it needs to at least double the size of features space. In the pairwise case, one-hot encoding is equivalent to the concatenation of features of two samples to generate the pairwise features. Considering the large expansion of training set by permutation, the further expansion in the feature size can greatly increase training time. Furthermore, our experiments on ChEMBL datasets have shown that one-hot encoding made little difference in the training accuracy. Therefore, we decided to use ordinal encoding for the pairwise features. In ordinal encoding, the choice of the integer value for each category is not restricted [20]. Despite potential doubts regarding the effect of their relative magnitudes under numeric transformations [21], it has been proven not to affect our study through simple tests. We endeavoured to assign each combination listed above with a different value (e.g., \({x}_{\mathrm{A},i}\) = 1, \({x}_{\mathrm{B},i}\)= 1 \(\to {X}_{\mathrm{AB},i}\) = -1; \({x}_{\mathrm{A},i}\) = 0, \({x}_{\mathrm{B},i}\)= 1 \(\to {X}_{\mathrm{AB},i}\) = 0). We have also tried a different set of ordinal values, for example, using {1, 2, 3, 4} instead of {-1, 0, 1, 2}. In both tests the results were hardly varied by the choice of ordinal values.
The pairwise target value needs to represent the difference in target values. For a specific pair, \({P}_{\mathrm{AB}}\), its pairwise target value, \({Y}_{\mathrm{AB}}\), is equal to \({y}_{\mathrm{A}}\) − \({y}_{\mathrm{B}}\). Pairs \({P}_{\mathrm{AB}}\) and \({P}_{\mathrm{BA}}\) are treated differently as two pairwise samples despite \({Y}_{\mathrm{AB}}\) = −\({Y}_{\mathrm{BA}}\). A ML method can learn to predict the real values of those pairwise differences \({Y}_{ }\) via regression or learn to predict the sign of the pairwise differences, \({\mathrm{sign}(Y) }\) via classification. The latter type of learning was found to be more advantageous to extrapolate the model and find extraordinary samples (see Sect. 2.4).
Suppose a dataset is split into a training set of size \({N}_{tr}\) and a test set of size \({N}_{ts}\). The training samples are paired via permutation, creating \({N}_{tr}^{2}\) pairwise training pairs. This type of pairs is referred to as C1-type training pairs in this study. The test pairs can be obtained in two ways: (1) C3-type test pairs: generate from a permutation of test samples, giving \({N}_{ts}^{2}\) test pairs; (2) C2-type test pairs: generate from pairing test samples with training samples, giving \(2{N}_{tr}{N}_{ts}\) test pairs. The naming of the pair types follows the notation in [22] which considers the amount of shared information between training and test data within a pair. Because this work is about the extrapolation of the pairwise approach, C2-type test pairs are more studied than the C3-type test pairs due to their ability to compare between training samples and test samples.
2.4 Extrapolation Strategy
The pairwise model only predicts the differences of pairs of samples. Therefore, a conclusive decision needs to be made to point out the predicted extraordinary samples. We propose to use rating algorithms to estimate the ranking of the test samples with the training set. The idea is to treat each predicted difference as the result of “a game match” between two samples. If the difference between sample A and sample B is greater than 0, then sample A wins sample B. This “league table of samples” gets updated from the predicted differences of the test pairs. In the end, we can identify the extraordinary or top-performing test samples.
Most of the generic rating algorithms were developed based on absolute wins or losses to give rating scores to the players, such as Elo’s rating algorithm for chess competition and Trueskill for computer games [23, 24]. There has also been some advanced research that enables these methods to take score differences to help the rating [25]. But for their application in the pairwise approach, it was found that the former version can serve our purpose better than the latter advanced version. We have noticed that if the pairwise model is trained on signed differences via classification, the accuracy of the predicted signs (wins, losses and draws) is higher than if it is trained on numerical differences via regression, from which the signs are then extracted. In other words, the accuracy of \({\mathrm{sign}(Y)}^{\mathrm{pred}}\) is higher than that of \({\mathrm{sign}(Y}^{\mathrm{pred}})\). This result may come from the fact that there exist pairs with the same differences in features (X) but different differences in target values (Y). Despite some loss of information when taking the signs, the training of the classification model can suffer from less “noise” in pairwise target values than the regression model. For a rating algorithm, correct results of win or loss are more informative in deciding the rank of the samples than the more accurate numerical score differences with potentially wrong signs. Therefore, training the pairwise model via classification and generic version of rating algorithm were used.
We have also experimentally examined several generic ranking algorithms and found that the choice of the generic ranking algorithm can merely affect the ranking accuracy given the same sets of \({\mathrm{sign}(Y)}^{\mathrm{pred}}\), usually by about 1%. It is believed that the main contribution to accurate ranking should come from the accuracy in \({\mathrm{sign}(Y)}^{\mathrm{pred}}\) rather than the rating algorithm. Therefore, Trueskill is selected and used to rank the samples from the predicted signs. Trueskill is originally designed to rank players in the game “Halo”. Because it assumes variances both in players’ performance and skill levels, it can deal with potential conflicts in match outcomes, in our case, conflicts in \({\mathrm{sign}(Y)}^{\mathrm{pred}}\) due to learning errors. For example, when \({\mathrm{sign}(Y)}_{AB }^{\mathrm{pred}}\)= –1 and \({\mathrm{sign}(Y)}_{BC }^{\mathrm{pred}}\)= –1, it implies that sample A < sample B < sample C. But if \({\mathrm{sign}(Y)}_{AC }^{\mathrm{pred}}\)= 1, which implies sample A > sample C, then these predictions are suggesting opposite opinions. This situation is similar to game tournaments, in which a strong player does not necessarily win every time. The python package for Trueskill was already made available [26]. In our experiment, the default Trueskill parameters were used.
In an extrapolation task, the relationship between the test samples and the training samples is important for comparing the training and test data in order to predict the extraordinary samples. So, despite the existence of C3-type test pairs, using them to rank solely can only tell the relative ranks within the test set. On the other hand, C2-type test pairs describe the relative differences between training and test samples. These are better suited for the extrapolation task. Therefore, in the following experiments on extrapolation, the signed differences of C2-type test pairs will be primarily used to rank.
2.5 Machine Learning Methods
The pairwise formulation is potentially ML method agnostic. We utilised the most common ML methods applied to QSAR learning: support vector machines (SVMs), random forests (RFs), Gradient Boosting Machine (XGBs) and K-nearest neighbours (KNN). We did not use deep learning as the datasets were generally too small. The ML methods used in this study are all based on the open-source ML python library, scikit-learn [27]. When a ML method is used to compare the standard and pairwise models, it is used with the default parameter setting from scikit-learn.
The pairwise approach uses classification for the predictions of signed differences, we therefore compared classification version of each ML method versus the standard regression approach. For each evaluation, 10-fold cross validation is used.
2.6 Extrapolation Metrics
To evaluate the extrapolation ability of a ML method, metrics other than the traditional evaluation metrics, such as mean squared error and R2, are required. This is because the common metrics are usually designed to cover predicted results over the whole test set, resulting in an averaged performance evaluation for both interpolation and extrapolation. In a random splitting in cross validation, the test set usually contains more interpolating samples than the extrapolating samples. Therefore, these metrics are good for evaluating the interpolation power of a model, but not very informative in terms of extrapolation power [6]. In this study, we decided to adopt the classification metrics of precision, recall and f1 score to count the identification of extraordinary and top-performing samples [1, 6]. This will give a more direct view of how useful a ML method is in an application where identifying top-performing samples is highly desired.
3 Results and Discussion
3.1 The Pairwise Approach Extrapolates Better
Our extrapolation experiments on 1436 ChEMBL datasets showed a clear advantage of the pairwise approach over the standard approach (Table 1 and Fig. 3). The ChEMBL datasets were sorted by size and experimented in order. When comparing the two approaches, the standard approach uses the regression version of a ML method to predict target values \(y\) and rank the test samples with training samples by predicted target values, while the pairwise approach uses the classification version of that ML method to predict \({\mathrm{sign}({Y}_{C2})}\) to rank the whole dataset.
It was found that the pairwise approach was much better at recognising the extraordinary and top-performing molecules than the standard approach. For all the three ML method (RF, SVM and XGB) tested, the pairwise approach almost always found equally or more extraordinary molecules than the standard approach (Table 1a). It can also identify more test molecules ranked within top 10% of the dataset most of the time, as shown by a high percentage for \({r}_{top10\%}\). Its outperformance in \({p}_{top10\%}\) is not as good as that in \({r}_{top10\%}\), but is still overall better than the standard approach. However, it was noted that this outperformance is less good for XGB or for larger datasets. This means that the ratio of false positives in the top-performing molecules by the pairwise approach can sometimes be similar to that by the standard approach. At the same time the pairwise approach often caused a greater increase in recall, which means it proposed more true positives. Hence, despite an outperformance in \({p}_{top10\%}\), the pairwise approach could propose slightly more false positives together with more true positives.
As extraordinary molecules do not necessarily exist every time when a train-test split is made, there were many datasets showing \({p}_{extra} = {r}_{extra}={f1}_{extra}=0\) or non-existing. Therefore, to illustrate outperformance, the datasets showing equal performance were removed. The percentage of datasets suggesting the pairwise approach outperformed the standard approach were re-calculated for the rest of the datasets (Table 1b). Across the three ML methods tested, the pairwise approach did outperform the standard approach in finding both the extraordinary and top-performing molecules. The results also suggested that using RF or XGB had less outperformance than SVM. Through further investigation, we found that the difference among ML methods was due to the variation in extrapolation performance by the standard approach. The standard approach using RF and XGB can evidently produce higher extrapolation metrics than using SVM for the ChEMBL datasets. At the same time, the pairwise approach performed similarly via both ML methods. This gives rise to the higher percentage of datasets showing pairwise approach was better with SVM in Table 1 and Fig. 3.
Apart from a statistical overview of the extrapolation power of the pairwise approach, we had a close look at its performance versus the size of the datasets. Figure 4 shows an example of the increase in \({f1}_{top10\%}\) versus the size of datasets for the experiments with RF for 1436 datasets of sizes from 30 to 298. The plots for other metrics showed a similar trend, that is the pairwise approach is more advantageous on smaller datasets, indicated by more data points above the line of ∆\({f1}_{top10\%}\)= 0 when the size of dataset is less than 200. This is mainly due to the standard approach learning better when the size of the dataset was larger, reducing the difference between the pairwise approach and the standard approach.
To test the generality of the paired formulation on other application datasets, we applied the same comparison experiment to a set of human gene expression datasets. Because each dataset contains 118050 rows of experimental conditions (samples), if the pairwise approach is applied for this size, the pairwise training set will be too large to train given any reasonable computational resources. We therefore decided to randomly sample a size 100 or 200 from each of the 978 gene datasets to compare the extrapolation performance. The extrapolation metrics were evaluated for the standard and the pairwise approach across four ML methods, random forest (RF), support vector machine (SVM), k-nearest neighbour (KNN) and gradient boosting machine (XGB).
We can see from Table 2 that for the gene expression datasets, the pairwise approach followed the trend seen in the ChEMBL experiments to outperform the standard approach. When the size of the datasets increased from 100 to 200, some of the extrapolation metrics decreased. This is also because the standard approach improved its learning through the additional data at a rate slightly greater than the pairwise approach, resulting in a decrease in the percentage of datasets showing outperformance. This is consistent with observations from Fig. 4.
3.2 The Extrapolation Strategy Improves Extrapolation
As shown in Sect. 2.4, we proposed a strategy to utilise the predictions of the pairwise models to give a ranking of training and test sets combined. This strategy is not exclusive to the pairwise approach. It can be applied to the standard approach to improve its extrapolation performance (Table 3). Once the standard approach has predicted the target values for the test set, the signed differences of C2-type or C3-type test pairs can be calculated from \({y}_{\mathrm{train}}^{\mathrm{true}}\) and \({y}_{\mathrm{test}}^{\mathrm{pred}}\). By inputting these signs to the rating algorithm, a ranking of the dataset can be obtained for further extrapolation evaluation. We will abbreviate the results from this procedure as the “standard rank approach”. Likewise, we compared the standard approach and the standard rank approach on ChEMBL datasets, which were sorted by size and experimented in order. Each of them was trained and tested via RF with 10-fold cross validation. In this experiment, we also compared the extrapolation results from both rankings obtained from C2-type test pairs and from C2-type test pairs plus C3-type test pairs.
F1 scores obtained by the pairwise approach (PA) or the standard rank approach (SRA) versus f1 scores obtained by the standard approach over 1456 ChEMBL datasets using RF. The upper row is the results from ranking with C2-type test pairs, whereas the lower row is from ranking with C2-type + C3-type test pairs.
The results in Table 3 show that the proposed extrapolation strategy can evidently enable the standard regression to identify more extraordinary samples compared to the direct regression with RF, which in theory is incapable to extrapolate outside the range of training targe values. By taking the signs and re-ranking the samples, despite at a cost of reducing the overall ranking correlation, which might have caused a reduced number of identified top-performing samples, Trueskill had the chance to re-allocate their relative positions by updating the probability distribution for each sample’s rating score. Because Trueskill updates the samples’ rating scores by numbers of pairwise comparisons, the more comparisons are entered the Trueskill algorithm, the more accurate and confident the rating scores will be. This might account for the increased number of datasets finding more top-performing molecules when C2-type and C3-type pairs are both used to rank. We also tested the case when C1-type training pairs, C2-type and C3-type test pairs are all entered the Trueskill and indeed the extrapolation performance was even better. To validate properly from a ML methodology standpoint the results are not included due to its use of training pairs.
The main differences that distinguish the standard rank approach from the pairwise approach are that (1) its calculated signed differences are all non-conflicting and consistent with each other, (2) its prediction objective focuses on the accuracy of predicted target values, and (3) the extraordinary samples are more likely to be predicted to draw (Y = 0) with the best training samples than to win (Y = 1) them. We found that the pairwise approach still can achieve a better extrapolation performance than the standard rank approach (see Fig. 5 and Table 4). This indicates that the pairwise model can produce a set of signed differences that better describes the relative positions of the training and test samples, resulting in the outperformance in extrapolation.
3.3 Discussion
The pairwise formulation is a method of combing model-reconfiguration and feature pre-processing techniques, rather than a new ML algorithm. It can be applied to multiple types of ML. The new formulation transforms the ML learning objective so that the emphasis is placed on the relationship between training and test samples. For a standard approach, when ML algorithms learn from seen examples and try to predict unseen examples from their “experience”, it can be difficult to extrapolate out of its “experience” domain. The pairwise approach, on the other hand, learns from the differences in features, which are sometimes more common and generalisable than the original features. It learns to predict the difference between training and test sample, directly aiming to predict if a test sample could win over the training samples. This transformed objective brings about the extrapolation performance of the pairwise formulation.
This study also recommends using classification metrics to evaluate extrapolation performance in a direct way. These metrics suit practical uses when extrapolation is required to identify the extraordinary samples from a test set. For example, they can be used to select ML algorithm for active learning. Active learning (AL) is a learning algorithm that interactively selects unlabelled samples to be labelled to learn the model in a goal-oriented way. In the selection, the exploration and exploitation are usually balanced so that AL can both improve the model’s applicability to a larger domain and improve the model’s prediction accuracy for the samples with desired properties. Hence, these extrapolation metrics can be used to assess and select ML methods with the desired exploitation property.
We believe that the extrapolation ability of the pairwise approach could be employed directly to fulfil the exploitation duty in an AL task for top-performing samples. Tynes et al. have also discovered the advantage of a pairwise approach for uncertainty-driven AL tasks, which encourages the exploration of the wider domain by selecting samples with less confident predictions [28]. We believe that it is possible to develop pairwise-approach-based AL, combining both the exploration and extrapolation traits found by Tynes’s study and ours. Despite the difference in how our pairwise approaches generate the pairwise features, it is ultimately the difference induced by data pre-processing techniques, which makes little differences between the two.
The main limitation of the pairwise approach is the additional time and memory requirement to train a pairwise model, as pointed out by Tynes et al. [28]. This is because the size of training set needs to be squared for the pairwise approach. Some techniques such as batch training and sub-sampling could certainly mitigate this. More generally, improvements in computer hardware will increasingly remove this limitation. Nevertheless, the pairwise approach can be useful in novel discovery projects with a limited budget or where data is scarce to better explore the surrounding space. In drug design, for example, accurate data points are expensive to generate, so it is important to utilise them efficiently. This study revealed the general applicability of the pairwise approach over thousands of datasets using default ML methods. Our next study will more thoroughly explore the new approach with tuned models on selected problems that demand extrapolation in order to mimic practical applications. To enable reproducibility, the code and datasets used for the experiments have been deposited on: https://anonymous.4open.science/r/pairwise_approach_extrapolation_2023-A188/
4 Conclusion
In this study we proposed a new pairwise configuration by first learning a classification function, F(sample1, sample 2) → signed difference in target values, then ranking the samples through Trueskill rating algorithm. We have compared for extrapolation the standard regression approach with our novel pairwise formulation. We found that the pairwise approach can almost always find more extraordinary samples from the test sets than the standard approach, across all the ML methods tested over 2400 ChEMBL and gene expression datasets. The pairwise approach outperformed the standard approach in identifying equally or more top-performing samples on ~ 70% of the datasets. It was also observed that the pairwise approach is more advantageous and effective when applied to smaller datasets. Additionally, we have found that this configuration can be adopted by the standard regression to identify more extraordinary samples. Yet the pairwise approach still outperformed the configured standard approach in all the extrapolation metrics tested.
References
Kauwe, S.K., Graser, J., Murdock, R., Sparks, T.D.: Can machine learning find extraordinary materials? Comput. Mater. Sci. 174, 109498 (2020). https://doi.org/10.1016/j.commatsci.2019.109498
Tong, W., Hong, H., **e, Q., Shi, L., Fang, H., Perkins, R.: Assessing QSAR Limitations – A Regulatory Perspective
Nicolotti, O. ed: Computational Toxicology: Methods and Protocols. Springer New York (2018). https://doi.org/10.1007/978-1-4939-7899-1
von Korff, M., Sander, T.: Limits of prediction for machine learning in drug discovery. Front. Pharmacol. 13, 832120 (2022). https://doi.org/10.3389/fphar.2022.832120
Cramer, R.D.: The inevitable QSAR renaissance. J. Comput. Aided Mol. Des. 26, 35–38 (2012). https://doi.org/10.1007/s10822-011-9495-0
**ong, Z., Cui, Y., Liu, Z., Zhao, Y., Hu, M., Hu, J.: Evaluating explorative prediction power of machine learning algorithms for materials discovery using k -fold forward cross-validation. Comput. Mater. Sci. 171, 109203 (2020). https://doi.org/10.1016/j.commatsci.2019.109203
Agarwal, S., Dugar, D., Sengupta, S.: Ranking chemical structures for drug discovery: a new machine learning approach. J. Chem. Inf. Model. 50, 716–731 (2010). https://doi.org/10.1021/ci9003865
Rathke, F., Hansen, K., Brefeld, U., Müller, K.-R.: StructRank: a new approach for ligand-based virtual screening. J. Chem. Inf. Model. 51, 83–92 (2011). https://doi.org/10.1021/ci100308f
Al-Dabbagh, M.M., Salim, N., Himmat, M., Ahmed, A., Saeed, F.: Quantum probability ranking principle for ligand-based virtual screening. J. Comput. Aided Mol. Des. 31, 365–378 (2017). https://doi.org/10.1007/s10822-016-0003-4
Liu, J., Ning, X.: Multi-assay-based compound prioritization via assistance utilization: a machine learning framework. J. Chem. Inf. Model. 57, 484–498 (2017). https://doi.org/10.1021/acs.jcim.6b00737
Zhang, W., et al.: When drug discovery meets web search: learning to rank for ligand-based virtual screening. J Cheminform. 7, 5 (2015). https://doi.org/10.1186/s13321-015-0052-z
Watson, O.P., Cortes-Ciriano, I., Taylor, A.R., Watson, J.A.: A decision-theoretic approach to the evaluation of machine learning algorithms in computational drug discovery. Bioinformatics 35, 4656–4663 (2019). https://doi.org/10.1093/bioinformatics/btz293
Meredig, B., et al.: Can machine learning identify the next high-temperature superconductor? Examining extrapolation performance for materials discovery. Mol. Syst. Des. Eng. 3, 819–825 (2018). https://doi.org/10.1039/C8ME00012C
King, R.D., Orhobor, O.I., Taylor, C.C.: Cross-validation is safe to use. Nat Mach Intell. 3, 276 (2021). https://doi.org/10.1038/s42256-021-00332-z
Mendez, D., et al.: ChEMBL: towards direct deposition of bioassay data. Nucleic Acids Res. 47, D930–D940 (2019). https://doi.org/10.1093/nar/gky1075
Olier, I., et al.: Transformational machine learning: Learning how to learn from many related scientific problems. Proc. Natl. Acad. Sci. U.S.A. 118, e2108013118 (2021). https://doi.org/10.1073/pnas.2108013118
Mayr, A., et al.: Large-scale comparison of machine learning methods for drug target prediction on ChEMBL. Chem. Sci. 9, 5441–5451 (2018). https://doi.org/10.1039/C8SC00148K
Morgan, H.L.: The generation of a unique machine description for chemical structures-a technique developed at chemical abstracts service. J. Chem. Doc. 5, 107–113 (1965). https://doi.org/10.1021/c160017a018
Koleti, A., et al.: Data portal for the library of integrated network-based cellular signatures (LINCS) program: integrated access to diverse large-scale cellular perturbation response data. Nucleic Acids Res. 46, D558–D566 (2018). https://doi.org/10.1093/nar/gkx1063
Brownlee, J.: Data Preparation for Machine Learning: Data Cleaning, Feature Selection, and Data Transforms in Python. Machine Learning Mastery (2020)
Kunanbayev, K., Temirbek, I., Zollanvari, A.: Complex encoding. In: 2021 International Joint Conference on Neural Networks (IJCNN), pp. 1–6. IEEE, Shenzhen, China (2021). https://doi.org/10.1109/IJCNN52387.2021.9534094
Park, Y., Marcotte, E.M.: Flaws in evaluation schemes for pair-input computational predictions. Nat. Methods 9, 1134–1136 (2012). https://doi.org/10.1038/nmeth.2259
Herbrich, R., Minka, T., Graepel, T.: TrueSkill(TM): A Bayesian skill rating system. In: Presented at the Advances in Neural Information Processing Systems 20 January 1 (2007)
Elo, A.E.: The Rating of Chessplayers, Past and Present. Arco Pub. (1978)
Hubáček, O., Šourek, G., železný, F.: Forty years of score-based soccer match outcome prediction: an experimental review. IMA J. Manage. Math. 33, 1–18 (2022)https://doi.org/10.1093/imaman/dpab029
TrueSkill — trueskill 0.4.5 documentation. https://trueskill.org/. Accessed 25 Apr 2023
Pedregosa, F., et al.: Scikit-learn: machine learning in python. J. Mach. Learn. Res. 12, 2825–2830 (2011)
Tynes, M., et al.: Pairwise difference regression: a machine learning meta-algorithm for improved prediction and uncertainty quantification in chemical search. J. Chem. Inf. Model. 61, 3846–3857 (2021). https://doi.org/10.1021/acs.jcim.1c00670
Acknowledgement
This work was partially supported by the Wallenberg AI, Autonomous Systems and Software Program (WASP) funded by the Alice Wallenberg Foundation. Funding was also provided by the Chalmers AI Research Centre and the UK Engineering and Physical Sciences Research Council (EPSRC) grant nos: EP/R022925/2 and EP/W004801/1. HN is supported by the EPSRC under the program grant EP/S026347/1 and the Alan Turing Institute under the EPSRC grant EP/N510129/1.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2023 The Author(s)
About this paper
Cite this paper
Wang, Y., King, R.D. (2023). Extrapolation is Not the Same as Interpolation. In: Bifet, A., Lorena, A.C., Ribeiro, R.P., Gama, J., Abreu, P.H. (eds) Discovery Science. DS 2023. Lecture Notes in Computer Science(), vol 14276. Springer, Cham. https://doi.org/10.1007/978-3-031-45275-8_19
Download citation
DOI: https://doi.org/10.1007/978-3-031-45275-8_19
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-45274-1
Online ISBN: 978-3-031-45275-8
eBook Packages: Computer ScienceComputer Science (R0)