Abstract
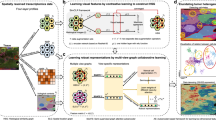
Lung adenocarcinoma has histologically distinct growth patterns that have been associated with patient prognosis. Precision segmentation of growth patterns in routine histology samples is challenging due to the complexity of patterns and high intra-class variability. In this paper, we present a novel model with a multi-stream architecture, Cross-Stream Interactions (CroSIn), which fully considers crucial interactions across scales to gather abundant information. The first-order attention introduces contextual information at an early stage to guide low-level feature encoding. The second-order attention then focuses on learning high-level feature relations among scales to extract discriminative features. Experimental results show interactions at both low- and high-level feature learning stages are crucial in performance improvement. The proposed method outperforms state-of-the-art networks, achieving an average Dice of \(60.34\%\) at patch level, and an average accuracy of \(65.31\%\) at sample level, which is also verified in an independent cohort.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
AbdulJabbar, K., Raza, S.E.A., Rosenthal, R., Jamal-Hanjani, M., Veeriah, S., et al.: Geospatial immune variability illuminates differential evolution of lung adenocarcinoma. Nat. Med. 26(7), 1054–1062 (2020)
Alsubaie, N., Shaban, M., Snead, D., Khurram, A., Rajpoot, N.: A multi-resolution deep learning framework for lung adenocarcinoma growth pattern classification. In: Nixon, M., Mahmoodi, S., Zwiggelaar, R. (eds.) MIUA 2018. CCIS, vol. 894, pp. 3–11. Springer, Cham (2018). https://doi.org/10.1007/978-3-319-95921-4_1
Chen, H., Qi, X., Yu, L., Heng, P.A.: Dcan: deep contour-aware networks for accurate gland segmentation. In: CVPR, pp. 2487–2496 (2016)
Chen, L.C., Yang, Y., Wang, J., Xu, W., Yuille, A.L.: Attention to scale: scale-aware semantic image segmentation. In: CVPR, pp. 3640–3649 (2016)
Chen, L.C., Zhu, Y., Papandreou, G., Schroff, F., Adam, H.: Encoder-decoder with atrous separable convolution for semantic image segmentation. In: ECCV, pp. 801–818 (2018)
Chen, S., Tan, X., Wang, B., Hu, X.: Reverse attention for salient object detection. In: ECCV, pp. 234–250 (2018)
Coudray, N., Ocampo, P.S., Sakellaropoulos, T., Narula, N., Snuderl, M., et al.: Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat. Med. 24(10), 1559–1567 (2018)
Feng, Z., Wang, Z., Wang, X., Mao, Y., Li, T., et al.: Mutual-complementing framework for nuclei detection and segmentation in pathology image. In: CVPR, pp. 4036–4045 (2021)
Fu, J., Liu, J., Tian, H., Li, Y., Bao, Y., et al.: Dual attention network for scene segmentation. In: CVPR, pp. 3146–3154 (2019)
Gertych, A., Swiderska-Chadaj, Z., Ma, Z., Ing, N., Markiewicz, T., et al.: Convolutional neural networks can accurately distinguish four histologic growth patterns of lung adenocarcinoma in digital slides. Sci. Rep. 9(1), 1–12 (2019)
Graham, S., Chen, H., Gamper, J., Dou, Q., Heng, P.A., et al.: Mild-net: minimal information loss dilated network for gland instance segmentation in colon histology images. Med. Image Anal. 52, 199–211 (2019)
Hashimoto, N., Fukushima, D., Koga, R., Takagi, Y., Ko, K., et al.: Multi-scale domain-adversarial multiple-instance CNN for cancer subtype classification with unannotated histopathological images. In: CVPR, pp. 3852–3861 (2020)
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: CVPR, pp. 770–778 (2016)
Huang, Q., **a, C., Wu, C., Li, S., Wang, Y., et al.: Semantic segmentation with reverse attention. In: BMVC (2017)
Kadota, K., Yeh, Y.C., Sima, C.S., Rusch, V.W., Moreira, A.L., et al.: The cribriform pattern identifies a subset of acinar predominant tumors with poor prognosis in patients with stage i lung adenocarcinoma: a conceptual proposal to classify cribriform predominant tumors as a distinct histologic subtype. Mod. Pathol. 27(5), 690–700 (2014)
Lin, T.Y., RoyChowdhury, A., Maji, S.: Bilinear CNN models for fine-grained visual recognition. In: ICCV, pp. 1449–1457 (2015)
Liu, D., Zhang, D., Song, Y., Zhang, F., O’Donnell, L., et al.: Unsupervised instance segmentation in microscopy images via panoptic domain adaptation and task re-weighting. In: CVPR, pp. 4243–4252 (2020)
Moore, D.A., et al.: In situ growth in early lung adenocarcinoma may represent precursor growth or invasive clone outgrowth-a clinically relevant distinction. Mod. Pathol. 32(8), 1095–1105 (2019)
Moreira, A.L., Ocampo, P.S., **a, Y., Zhong, H., Russell, P.A., et al.: A grading system for invasive pulmonary adenocarcinoma: a proposal from the international association for the study of lung cancer pathology committee. J. Thorac. Oncol. 15(10), 1599–1610 (2020)
Oktay, O., Schlemper, J., Folgoc, L.L., Lee, M., Heinrich, M., et al.: Attention u-net: learning where to look for the pancreas. In: MIDL (2018)
Qaiser, T., Tsang, Y.W., Taniyama, D., Sakamoto, N., Nakane, K., et al.: Fast and accurate tumor segmentation of histology images using persistent homology and deep convolutional features. Med. Image Anal. 55, 1–14 (2019)
Sirinukunwattana, K., Pluim, J.P.W., Chen, H., Qi, X., Heng, P., et al.: Gland segmentation in colon histology images: the glas challenge contest. Med. Image Anal. 35, 489–502 (2017)
Tokunaga, H., Iwana, B.K., Teramoto, Y., Yoshizawa, A., Bise, R.: Negative pseudo labeling using class proportion for semantic segmentation in pathology. In: Vedaldi, A., Bischof, H., Brox, T., Frahm, J.-M. (eds.) ECCV 2020. LNCS, vol. 12360, pp. 430–446. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-58555-6_26
Tokunaga, H., Teramoto, Y., Yoshizawa, A., Bise, R.: Adaptive weighting multi-field-of-view CNN for semantic segmentation in pathology. In: CVPR, pp. 12597–12606 (2019)
Travis, W.D., Brambilla, E., Nicholson, A.G., Yatabe, Y., Austin, J.H., et al.: The 2015 world health organization classification of lung tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J. Thorac. Oncol. 10(9), 1243–1260 (2015)
Valanarasu, J.M.J., Oza, P., Hacihaliloglu, I., Patel, V.M.: Medical transformer: gated axial-attention for medical image segmentation. In: MICCAI (2021)
Wang, X., Fang, Y., Yang, S., Zhu, D., Wang, M., et al.: A hybrid network for automatic hepatocellular carcinoma segmentation in H &E-stained whole slide images. Med. Image Anal. 68, 101914 (2021)
Warth, A., Muley, T., Kossakowski, C., Stenzinger, A., Schirmacher, P., et al.: Prognostic impact and clinicopathological correlations of the cribriform pattern in pulmonary adenocarcinoma. J. Thorac. Oncol. 10(4), 638–644 (2015)
Wei, J.W., Tafe, L.J., Linnik, Y.A., Vaickus, L.J., Tomita, N., Hassanpour, S.: Pathologist-level classification of histologic patterns on resected lung adenocarcinoma slides with deep neural networks. Sci. Rep. 9(1), 1–8 (2019)
Woo, S., Park, J., Lee, J.Y., Kweon, I.S.: CBAM: convolutional block attention module. In: ECCV, pp. 3–19 (2018)
Yu, C., Zhao, X., Zheng, Q., Zhang, P., You, X.: Hierarchical bilinear pooling for fine-grained visual recognition. In: ECCV, pp. 574–589 (2018)
Zhao, H., Shi, J., Qi, X., Wang, X., Jia, J.: Pyramid scene parsing network. In: CVPR, pp. 2881–2890 (2017)
Zhou, Y., Chen, H., Xu, J., Dou, Q., Heng, P.-A.: IRNet: instance relation network for overlap** cervical cell segmentation. In: Shen, D., et al. (eds.) MICCAI 2019. LNCS, vol. 11764, pp. 640–648. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-32239-7_71
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Appendix
Appendix
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Pan, X. et al. (2022). Cross-Stream Interactions: Segmentation of Lung Adenocarcinoma Growth Patterns. In: Qin, W., Zaki, N., Zhang, F., Wu, J., Yang, F. (eds) Computational Mathematics Modeling in Cancer Analysis. CMMCA 2022. Lecture Notes in Computer Science, vol 13574. Springer, Cham. https://doi.org/10.1007/978-3-031-17266-3_8
Download citation
DOI: https://doi.org/10.1007/978-3-031-17266-3_8
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-17265-6
Online ISBN: 978-3-031-17266-3
eBook Packages: Computer ScienceComputer Science (R0)