Abstract
In the 1970s, the number of accessions held in national and international seed and gene banks increased steadily. This growth, initially a source of pride, was recognised as a liability by the 1980s. Too many accessions lacked the basic information necessary for researchers to access and use samples knowledgably. Many gene banks came under scrutiny for poor management practices and several found themselves accused of mishandling a ‘global patrimony’ entrusted to their care. In this paper, I explore one response to these concerns that attracted attention from many in the germplasm conservation community: creating linked, standardised databases of collections. Calls for more and better data about accessions often emphasised that these data would make collections easier to use and therefore more valued. Here I take a close look at the early history of data collation and standardisation as a means of ‘rationalising’ collections, a motivation that was not advertised as prominently. This historical example shows the infrastructures developed to facilitate data exchange in the context of seed and gene banking to have been tied up with both mundane imperatives to cut costs and lofty goals of building political bridges—in addition to the often-repeated ambition of making plant breeding more efficient.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
1 Introduction
In 2010, the United Nations Food and Agriculture Organisation (FAO) released its Second Report on the State of the World’s Plant Genetic Resources for Food and Agriculture, a status update on seed and gene bank collections and other activities aimed at conserving crop diversity worldwide. Based on accounts received from national, regional, and international institutions, the Second Report estimated that, collectively, the world’s seed and gene banks maintained some 7.4 million accessions (that is, individual samples registered in collections), a 20% increase over the preceding 20 years (FAO, 2010). From the perspective of preserving examples of farmers’ varieties and crop wild relatives—conceived since the 1970s as vanishing ‘genetic resources’, but recognised much earlier as potentially endangered by breeders’ creations—the continued growth of collections would seem cause for celebration (Fenzi & Bonneuil, 2016; Bonneuil, 2019). Yet the compilers of the Second Report cautioned against the easy interpretation of growth as victory. As they noted, most of this expansion had not come from acquiring new materials in the field, despite the fact that many minor crops and wild relatives desperately needed such attention. The impressive increase in collections was instead ‘the result of exchange and unplanned duplication’ of existing accessions. Scientists’ and administrators’ concerns about the implications of undirected growth can be read into the conclusion to their opening summary statement: ‘There is still a need for greater rationalization among collections globally’ (FAO, 2010: xix).
This was not a new demand. As I describe in this paper, calls to ‘rationalise’ gene banks, especially though not exclusively through the elimination of duplicate materials, date to the late 1970s. That decade saw a steady increase in the numbers of accessions held in national and international collections, expansion that was driven by concern about rapid ‘genetic erosion’ in the wake of the Green Revolution and perceived widespread agricultural industrialisation. The quick expansion of collections, initially a source of pride, was by the end of the decade recognised as a liability. Too many accessions lacked the basic information necessary for researchers to make requests of gene bank managers, let alone put samples to work knowledgably in breeding programmes. In the early 1980s, many gene banks came under scrutiny for poor management practices, and several prominent banks found themselves accused of mishandling a ‘global patrimony’ entrusted to them by the international community. One response to these failings, real and perceived, attracted attention from many in the germplasm conservation community: creating linked, standardised databases of collections. Calls for more thorough and consistent data about accessions often emphasised, and still emphasise today, that these data will make collections easier to navigate and therefore more valued and more used (e.g. Weise et al., 2020).
In this chapter I take a close look at the early history of data collation and standardisation as a means of ‘rationalising’ gene bank collections, a motivation that was not advertised as prominently. For some researchers and collection managers, the identification of duplicates was thought to allow the channelling of limited time and money to only the most unique accessions, even creating the possibility of de-accessioning items known to be held elsewhere. My analysis calls attention to three elements of this history in particular. First, I note the diverging functions of evaluation data and other identifying information within seed and gene banks in the late 1970s and 1980s, when these were sought both to encourage greater use of collections (e.g. in breeding programmes) and also to better manage collections (e.g. to eliminate duplicates). Second, I examine the political motivations that lay behind some calls for rationalisation. Where rationalisation was to include the elimination of duplicates across gene banks, it promised to save precious time and money and also to forge trust and interdependence among politically divided scientists, institutions, and states. Third, I explore how the ability of rationalisation initiatives to meet either economic or political objectives was frustrated by technical hurdles in data management, limited personnel, financial constraints, and political obstacles.
Ultimately this historical example shows the infrastructures developed to facilitate data exchange in the context of seed and gene banking to have been tied up with mundane imperatives to cut costs and lofty goals of building political bridges—in addition to the often-repeated ambition of making plant breeding more efficient and effective. By following imperatives issued from above out into the ‘field’ where curators wrestled with the chaos of actual collections, I show that the technical magic bullet of database development demanded, rather than generated, economic and political resources.
The political and economic imperatives behind data sharing are often neglected in historians’ and sociologists’ assessments of data practices associated with gene banks, which tend to focus on actors’ interest in deriving value from collections (e.g. Parry, 2004; Van Dooren, 2010; Fullilove, 2018; for an exception see Chacko, 2019). Over time, calls for more and better data (and better data curation, too) in and across gene banks have become entwined with even more ambitious data enterprises that seek to unify a vast array of information about crop germplasm (for examples see, in this volume, chapters by Harrison and Caccamo; Arnaud et al. and Devare, Arnaud and King). Unpicking the many competing factors—social, political, technical—that informed and impeded earlier efforts to build comprehensive data infrastructures may not only provide a richer historical picture, but also help today’s data developers recognise and navigate the complexities of their own present and future work.
2 Seed Surfeits, Data Shortfalls and the Call for Rationalisation
Much of today’s international infrastructure for the conservation of crop diversity—the breeders’ and farmers’ varieties and crop wild relatives collectively designated as ‘plant genetic resources for food and agriculture’—was forged in the 1970s and 1980s. Over the preceding century, plant breeders and other agricultural experts had called with increasing urgency for cross-border coordination of efforts to conserve crop genetic diversity (Lehmann, 1981; Bonneuil, 2019). As a number of historians have described, it was the real and perceived effects of international agricultural aid programmes of the late 1950s and 1960s (e.g., the ‘Green Revolution’) that finally galvanised international initiatives. These centred on collecting and arranging for long-term storage of breeders’ varieties, landraces, and crop wild relatives thought to be endangered (Pistorius, 1997; Fenzi & Bonneuil, 2016; Curry, 2017, 2022; Bonneuil, 2019). They also entailed further coordination efforts focused on data generation, systematisation, and exchange. Better data and more thorough data linkage were considered essential to making collections useful, manageable, accessible and cost effective (Curry & Leonelli, Forthcoming). However, as I describe here, it proved far easier to acquire samples than data about these, circumstances that drove and, paradoxically, frustrated calls for collection ‘rationalisation’.
From 1974, an International Board for Plant Genetic Resources (IBPGR), organised under the Consultative Group on International Agricultural Research (CGIAR), attempted to coordinate the conservation efforts of national institutions and international agricultural research centres and to encourage further programmes through strategic sponsorship of collecting missions and conservation facilities (Curry, 2017). It also aspired to establish a network of ‘base collections’, which would link national and international gene banks with especially good infrastructure and management (Hanson et al., 1984; Thormann et al., 2019). This network would disperse the responsibility for administering an international gene bank—something that had been sought by many individuals and institutions in the preceding decades—across multiple sites. With administrators focused especially on technical capacities such as reliable temperature and humidity control, those sites ended up being the comparatively well-resourced genetic resources programmes of industrialised countries and the internationally financed agricultural research centres of the CGIAR (Peres, 2019).
By the mid-1980s, these international coordinating efforts had fostered hundreds of collecting missions and many new conservation programmes. A 1984 tally estimated that the IBPGR had been ‘instrumental in fielding over 300 collecting missions’ in 88 countries and in its first decade had placed over 100,000 samples in gene banks (Williams, 1984: 7). It had also come under intense scrutiny, in part because of its perceived effectiveness in securing seeds. In his influential 1979 book Seeds of the Earth, the Canadian activist Pat Mooney linked IBPGR sponsorship of collecting missions and its ferrying of seeds to well-resourced facilities in the United States and Europe (or to CGIAR institutions located in the Global South but managed largely from the North) to a long history of imperial exploitation. In Mooney’s assessment, ‘The emerging network of gene banks takes national genetic treasures from the Third World to be stored abroad. In effect, these national resources cross a technological frontier, robbing the world’s original plant breeders—subsistence farmers—of their rightful heritage, and leaving Third World governments dependent upon the First World for their own germplasm’ (Mooney, 1979: 102). Thanks in part to Mooney and a growing number of seed activists, the 1980s saw a powerful surge in critiques of, and resistance to, the international network of seed and gene bank facilities sought by IBPGR and its funders. These critiques eventually forced the reimagining of this global network (Aoki, 2008; Fenzi & Bonneuil, 2016).
Among other outcomes, the fight over control of seed fostered by activists like Mooney and pursued by the nonaligned states at FAO from the 1980s onward brought new scrutiny to seed banks (Fenzi, Forthcoming). Scientists and administrators associated with IBPGR needed to provide evidence that their work had been in the global interest and that its network of base collections was indeed kee** seeds safe and accessible to all potential users. Critics, meanwhile, needed proof of the opposite. Subsequent studies compiled many shortcomings of national and international conservation efforts: broken refrigeration systems, lost samples, restrictions on access (e.g., US Comptroller General, 1981; Goodman, 1984; Mooney, 1983). Even champions of the existing structures had to acknowledge that the putative success of gathering seed samples had created a significant influx of materials to conserve, and that this multiplied the labour needed in processing, monitoring, and evaluating samples (e.g., Frankel, 1984; Peeters & Williams, 1984). What’s more, the burgeoning size of collections had not been accompanied by increasing demand. A 1984 study of seed and gene bank use conducted by IBPGR, and co-authored by its executive secretary, described ‘a consensus of opinion that genebanks are not being used very extensively by breeders’ (Peeters & Williams, 1984: 22).
The acknowledgment that seed and gene banks often struggled to stay abreast of maintenance and almost always failed to provide meaningful services to breeders prompted calls for new strategies in conservation. A demand for more and better information about gene bank accessions—that is, for good data—featured centrally in many of these calls. In a clear signal of change, Otto Frankel, an early and effective champion of urgent collecting missions and institution building in the 1960s and 1970s, now advised a slowed pace for these. Frankel thought the use of collections was hampered by the lack of information about individual accessions, especially their agronomic traits and how they might be expected to perform in different environments (Frankel, 1984). This was not idle speculation. In 1984, IBPGR estimated that 95% of samples in gene banks had no such evaluation data attached (Peeters & Williams, 1984: 24). This was despite the fact that IBPGR had, since its founding, emphasised the creation, standardisation and computerisation of such data (Curry & Leonelli, Forthcoming).
By the mid-1980s, the imbalance between the cascade of collections and the dribble of data to accompany these—and the mounting critiques of its work—led the IBPGR to articulate a change in policy, a ‘period of consolidation’ in which ‘characterisation, documentation and the ready exchange of information’ would predominate (Williams, 1984: 14). One of the chief obstacles to this vision was that few if any people had capacity to systematically generate data. Initially, the IBPGR had assumed that national and international agricultural institutes would create this essential information, for example by carrying out evaluation programmes to generate data for individual accessions on agronomic qualities and environmental adaptations. However, although ‘it was thought [at IBPGR] that characterisation and preliminary evaluation would not be costly’ this initial view was quickly revised. Delays in generating these data were intensified by funding and staff shortfalls at many national programmes. In addition, the assumption that breeders would contribute to gene bank work by submitting any data they produced about requested accessions had to be scrapped. As the executive secretary of IBPGR bluntly summarised, ‘[B]reeders have not been very forthcoming in offering their services in this respect’ (Williams, 1984: 11). This was not necessarily a product of intransigence on the part of breeders. On the contrary, there simply were not many rewards to their spending time and energy returning information to gene banks about samples they studied. Even if they had done so, the data would likely have arrived in heterogenous forms, requiring further labour from curators, who along with breeders would also have been navigating the changing international standards for the crop descriptors scientists were exhorted to use (Curry & Leonelli, Forthcoming).
The labour- and resource-intensive nature of evaluation was complicated by another issue increasingly recognised as characteristic of the international conservation system: duplication. A 1984 study of seed and gene bank conservation based on a survey of some 760 scientists determined that ‘[a]t least 50% of the combined collections of most crop species are duplicate accessions’ (Lyman, 1984: 5). In some ways, this was a definite advantage. Having extra copies meant that disruptions at one seed or gene bank need not cause undue alarm. It also presented a problem, particularly given the finite nature of resources. ‘[I]ndiscriminate duplication of entire collections at numerous genebanks is costly and unnecessary’, the scientist preparing the report insisted, noting that ‘[r]edundant duplicates within the same bank are undesirable’ (Lyman, 1984: 5). If one were to add the labour of evaluating accessions to that of maintaining them, the unintended costs of accession duplication would only intensify. A conundrum followed, however: Unnecessary duplication increased the costs of maintaining collections, including the costs of evaluating these, but evaluation was also needed to identify duplicates if collections were to be rationalised (Lyman, 1984: 17).
Otto Frankel thought it was absurd to expect that seed and gene banks, with their limited resources, would be able to produce evaluation data for the thousands of samples they now maintained. His proposal was instead the ‘rationalization of evaluation’ (emphasis mine) through the selection of a ‘core collection’ of samples that were thought to represent most of the genetic diversity in the collection. This would entail ‘a drastic reduction in redundancy’, at least in terms of genetic variation within the identified core, and therefore also reduce the energy devoted to evaluation (Frankel, 1984: 161; see also Brown, 1989, 1995). The ‘rationalization of evaluation’ through the use of core collections found influential champions in the 1980s and 1990s (e.g. Brown & Spillane, 1999). A chief selling point of core collections was not that they would reduce the overall number of samples, but instead that they would ensure that the widest possible range of genetic diversity would be maintained and used even in circumstances of constrained resources. In fact, the core collection concept appealed precisely because it meant that streamlined gene bank management could occur without a costly investment in eliminating duplicate samples via field evaluations of an entire collection or even by newly available biochemical and molecular techniques. In the 1980s and 1990s, these were, for the most part, prohibitively expensive to run for entire seed bank collections. As a committee of experts assembled under the aegis of the US National Research Council acknowledged in 1991, ‘Elimination of redundancy in existing collections is not cost-effective’ (NRC, 1993: 172).
3 The Techno-political Project of Collection Decentralization
The merits of eliminating duplicates were, as the foregoing discussion suggests, often debated by gene bank managers and other experts in technical and economic terms. Scientists advocated approaches that they felt would produce the best conservation outcomes at the least expense. Yet the drive for rationalisation through data generation and de-duplication was at times a product of political considerations as much as technical ones—even beyond the unquestionably political project of showing that the ‘patrimony of humanity’ was well and affordably cared for in seed banks. A European Cooperative Programme on the Conservation and Exchange of Crop Genetic Resources (ECP/GR), which first took shape in the late 1970s, imagined the elimination of duplicates across collections as a crucial step in the creation of a decentralised European gene bank. As I discuss here, realising such a bank would depend not only on better data, but also on strong ties and mutual confidence among institutions as well as among the governments that sponsored those institutions. In this context, rationalisation through the elimination of duplicates was a project that depended on existing geopolitical relationships—and attempted to forge new ones.
The initial conversations that led to the ECP/GR took place in 1975. Although this timeframe—in sync with the founding of the International Board for Plant Genetic Resources—points to the influence of international mobilisations on this European project, the origins of the ECP/GR lay in regional, not global, concerns. As early planning documents described, the initiative was imagined within the UNDP’s European Office as contributing to that organisation’s ‘endeavour to establish cooperation between East and West European countries’ (FAO, 1979: 1). Thanks to early imperial infrastructures for acquiring and maintaining plant materials from around the world (Brockway, 1979; Drayton, 2000) and increasing state emphases on strategic collections of crop diversity from the 1920s onward (Pistorius & van Wijk, 1997; Flitner, 2003; Saraiva, 2013; Bonneuil, 2019), European institutions collectively possessed an estimated two-thirds of the world’s crop gene bank accessions (FAO, 1979: 15). The European Association for Research on Plant Breeding (EUCARPIA) had begun to link the activities of these institutions through its gene bank committee in 1966, focusing especially on coordinating collecting missions and agronomic characterisation of accessions. The planned ECP/GR would expand and deepen this coordination effort, with the aim of ‘permitting direct access on the part of every plant breeder to the germplasm of the entire continent… thus making possible a previously unattainable level of plant breeding efficiency’ (FAO, 1979: 17). Communication and harmonisation across European agricultural research organisations would benefit all breeders, and all nations, that participated. This could not be achieved simply through professional researchers and breeders acting independently out of their ‘somewhat limited goodwill’: it demanded the formal commitment of governments (FAO, 1980: 11).
This bridging of East and West to the benefit of all Europeans—and the ‘Third World’, too, as many planning documents insisted that shoring up the foundations of European gene banks would ramify well beyond the continent—would depend especially on generating data about accessions and ensuring that both data and the systems used to record these were in reasonable harmony. In the most general terms, ‘a major effort to describe and document all existing genetic resources collections in Europe’ would be accompanied by an ‘all-European genetic data exchange’, the latter produced by finding various means of making diverse existing gene bank data management systems interoperable (FAO, 1979: 18–19).
Achieving this level of data exchange would not just make accessions more readily accessible to researchers. It was also imagined as a route to reducing duplication: duplication of collecting missions, duplication of evaluation and characterisation programs, duplication of accessions themselves, and—of course—duplication of the expenditures needed to conduct any of these activities (FAO, 1979: 20; FAO, 1980: 3). For example, ‘The burden of collating comprehensive information about the genetic resources of crop plants could be shared between genebanks by each one accepting responsibility for the in-depth study of a particular crop (or crops)’ and making the results available to all other collections (ECP/GR, 1981a: 25). Having converged on these objectives, more than 20 European countries agreed to launch the ECP/GR in 1980, with start-up funding from UNDP and administrative support from the United Nations Food and Agriculture Organisation (FAO).Footnote 1
The programme’s governing body met for the first time in December 1980. Observers to the initial meeting and other early convenings of this body included nations unwilling to become full participants but interested in the proceedings (most notably, the Soviet Union) as well as organisations with relevant expertise, resources or both. The International Board for Plant Genetic Resources was an obvious collaborator, and its international mandate was seen as the route for delivering the promised payoffs of harmonisation within Europe to the wider world, especially agricultural research programmes in develo** countries. The IBPGR’s still nascent understanding of network-building at a global scale was complemented by the input of several organisations with network-building expertise at a sub-regional level: the Nordic Gene Bank, the IBPGR’s recently established Mediterranean Programme, and the genetic resources networks of both the European Economic Community and the Council for Mutual Economic Assistance (also known as COMECON) (FAO, 1979: 2; see also participant lists in meeting reports, e.g., ECP/GR, 1981a, b). These sub-regional groups had been established to facilitate precisely the type of coordination and exchange now imagined as a pan-European project. In a way, the European Cooperative Programme sought to knit together existing but geopolitically divided networks of researchers and institutions.
One way to forge lasting links was to create interdependence, unifying institutions by dividing labour. Although national representatives and other participants in the ECP/GR programme wanted ideally to establish ‘one or a few centralized genebanks in Europe’ they knew that the resources and will for creating new transnational institutions was in short supply. They therefore initially imagined existing institutions becoming ‘lead centres’ for a certain crop or several crops, taking on the responsibility for maintaining and providing access to all the accessions of that crop on behalf of participating countries. For example, an early list suggested the Plant Breeding and Acclimatization Institute in Radizkow, Poland would take responsibility for Secale (ryes), the National Vegetable Research Station in Wellesbourne, UK as the lead centre for Allium species (onions, garlic, leeks, etc.), and so on (ECP/GR, 1981a: 30–33; see also ECP/GR, 1981b: 38–44). This arrangement was thought to potentially economise on time and labour and—perhaps just as importantly—‘create mutual interest and build up confidence by making countries and sub-regions mutually dependent’ (ECP/GR, 1981a: 30).
Ultimately this vision of a decentralised European gene bank, to be created by networking specialised crop centres, gave way to a still more decentralised vision in which even the European crop collections would be generated by networking among different national collections rather than transferring responsibility to a single lead centre. With crop species as the definitive means of organising the network—rather than, say, ecological zones, regional boundaries, or working languages—the ECP/GR established ‘crop committees’ (later, ‘crop working groups’) for the crops its scientific advisors deemed most important to collect and conserve. An initial selection of 12 crops was based on criteria that included evolutionary history (European indigeneity), biocultural factors (significant genetic diversity in European landraces, unique national appreciation), economic and agronomic importance, and technical considerations (state of existing collections, quality of existing data) (ECP/GR, 1982: 25–26). This list was then narrowed to just six: barley, forages, Prunus (plums, cherries, peaches, almonds, etc.), Allium, oat, and sunflower. A handful of experts on the selected crops, representing institutions with significant existing collections of these, constituted the working groups. Their main objective was to find means of actualising the overarching ECP/GR goal of enhancing cooperation and reducing duplication in specific projects (UNDP-IBPGR, 1984: 4).
As the ECP/GR moved toward implementation, after multiple years of negotiation and planning, data generation and data management took centre-stage. Ensuring cross-institution ‘interoperability’ of data about collections had been seen from the outset as a crucial mechanism for cross-country coordination. But emphasis on this aspect was likely heightened by the decision to fold ECP/GR into the work of IBPGR in 1983. In 1981, while planning was still in progress, the executive secretary of IBPGR, J. Trevor Williams, had exhorted ECP/GR participants not to delay on what he saw as the most crucial element of more effective gene bank coordination: generating data. He insisted that ‘immediate action’ was needed to ‘put into order most of the collections by incorporating basic information into data bases’. These could then be used ‘to sort out redundant duplicates thereby leading to the maintenance of perhaps smaller, but well documented and more useful, collections’. He was particularly concerned that the group didn’t have a grasp of the true number of accessions it needed to manage, given the amount of ‘redundant duplication’ within and across institutions (ECP/GR, 1981b: 9–10).
Williams’ concerns about the problems of European collections reflected issues that IBPGR was grappling with more generally in the early 1980s. As discussed above, these included especially the significant uptick in collecting of the 1970s arising from increased attention to genetic erosion as a conservation issue, a subsequent expansion in collections, and the vulnerabilities in collections management generated as a result. Given the extent to which the world’s collections were in European hands, the IBPGR problem of seed surfeits and missing data could be understood as a largely European problem.
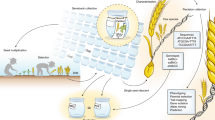
The emphasis on data and databases as essential and often neglected instruments for gene bank management contributed to the positioning of crop databases—information infrastructures containing ‘all information on the germplasm of each crop kept in European genebanks’ and maintained on a computer at a single leading institute—as a top priority for the crop working groups (ECP/GR, 1981b: Appendix VIII; UNDP-IBPGR, 1984: 9–15). These databases would be the chief tools by which the working groups (and by extension the ECP/GR) would coordinate conservation activities across Europe. They would be useful for not only rationalising collections by removing unwanted duplicates and moving towards decentralisation of collections, but also identifying gaps in European holdings that could be resolved through collecting missions and enabling strategic planning for evaluation and characterisation (UNDP-IBPGR, 1984). (See Fig. 1).
In 1984, the ECP/GR imagined the implementation of crop databases using the example of Allium. ‘Country coordinators’ would ensure that basic passport data existed for all national collections. These data would be fed into a European catalogue (i.e. the crop database) via manual or computerised questionnaires. Once all data were registered and the catalogue was complete, the latter would be the basis for rationalisation of collections and further activities including collecting, training, and characterisation/evaluation. (From UNDP-IBPGR, 1984, pp. 10–11). Reprinted by permission of Bioversity International
The general steps outlined for the crop working groups included, first, the compilation of accession lists for all of the samples of the relevant crop (and in some cases its wild relatives) in the gene banks of participating institutions. A second step was agreeing and implementing descriptors, that is, deciding on a consistent set of information to be associated with each accession and a consistent way of expressing that information. With complete accession lists in hand providing ‘basic passport descriptors’ (as opposed to more detailed evaluation or characterisation descriptors), working groups would be in a position to complete the third initial task, the identification of duplications. Coordination of further characterisation activities, with an eye to populating the database with still more useful agronomic data, would follow—or so the idealised workflow suggested (UNDP-IBPGR, 1984; Perret, 1985). In sum, European crop databases, created by asking experts from different institutions and nations to create, harmonise and pool data about existing crop gene bank collections, was the technical tool through which the political project of uniting European genetic resources—and by extension European scientists and governments—would be achieved.
4 Databases and De-duplication
The ECP/GR’s Barley Working Group made impressive progress towards the goals of develo** a database and deploying this to reduce duplication within and across collections in the programme’s first two decades. Its impacts nonetheless fell far short of those projected at the outset, consisting mostly in investigating the tools necessary for accomplishing decentralisation through de-duplication. A close look at the working group’s efforts in this period reveals the significant technical hurdles that database creation and decentralisation entailed. It also reveals the strategies that scientists and gene bank managers adopted in attempting to navigate the paradoxical situation outlined above—namely, that preventing costly duplication of data-generating evaluation programmes (along with other expenditures related to the maintenance of accessions) nonetheless depended on undertaking potentially costly data production and management exercises.
The ECP/GR prioritized barley early on in its proceedings. Together with forages and Prunus, barley was one of three crops considered important across all four sub-regions and inadequately addressed in other international programmes (ECP/GR, 1982: 25). The working group’s initial tally of barley accessions in ‘significant’ European gene banks suggested that there were about 85,000 of these—and that at least 60% represented samples duplicated across collections. The institution among participating nations with the largest number of barley accessions (about 9400) was the Zentralinstitut für Genetik und Kulturpflanzenforschung at Gatersleben, German Democratic Republic. This was subsequently designated the lead centre for barley conservation efforts and hosted the working group’s first meetings (Barley Working Group, 1983).Footnote 2 For the Barley Working Group, as for the other crop groups, the initial priority task was to develop a ‘European data base’ of all existing collections. This would come to be considered the ‘backbone of the work of the group’ (Dirk & Knüpffer, 2001: 50).
The Barley Working Group first outlined its plan for fulfilling its assigned tasks at a 1983 meeting in Gatersleben. Six members plus a chair formed the official working group, which hailed from institutions in Austria, Czechoslovakia, Denmark, the German Democratic Republic, the Netherlands, and Poland. This group elaborated a set of aims that hewed closely to the mandate it had been given. Top** its list of action items was ‘the complete documentation of European barley collections’ according to a standard list of descriptors, followed by ‘the registration of this data in computer data bases’, and the ‘detection of replication of accessions’. These would make possible ‘the rationalization of collections by agreement between participating gene banks with consequent elimination of potential waste of resources in the storage, multiplication, characterization and evaluation of redundant accessions’ (Barley Working Group, 1983: 1). In other words, immediate improvements in data creation, management and exchange would reduce the costs of collection management—including further costly data generation (i.e. evaluation of duplicate accessions). Documentation, database development and de-duplication would also result in a decentralised European barley gene bank managed not by any one institution but by all.
The first version of the European Barley Database, assembled between 1984 and 1987, brought together the passport data associated with over 55,000 barley accessions from more than 30 European collections in 26 countries. The database was maintained in Gatersleben at the Zentralinstitut für Genetik und Kulturpflanzenforschung on an 8-bit microcomputer (Knüpffer, 1988a, b; Dirk & Knüpffer, 2001: 50). Participants in the initial 1983 meeting of the Barley Working Group had been asked to bring with them information—‘if possible, computer printouts’—reporting on the contents of collections in their home country and neighbouring countries. Only four institutions provided the requested printouts, suggesting the extent to which most information regarding collections remained in records maintained chiefly by hand (Barley Working Group, 1983: 2) or otherwise difficult to share. The development of the database at Gatersleben was therefore a staged process requiring the acquisition of collection data from across institutions, the ‘preprocessing’ of these by a colleague in Sweden, the standardisation of the descriptors used in different data sets, and their eventual merger into a complete list of accessions maintained at Gatersleben (Knüpffer, 1988a, b).
With a first iteration of the database in hand in 1987, it was time to put it to use in coordinating across institutions, as imagined. As the database’s chief developer noted, it was already possible to send inquires and requests to Gatersleben such as ‘Print a list of all two-rowed winter barleys originating from the Far East’ or ‘Where could I ask for living seeds of the cultivars and strains listed below?’ and receive a reply from the scientist in charge of the database (Knüpffer, 1988b: 19) But this service provision was hardly the reason the database had been developed. Duplication remained a key concern of the Barley Working Group; by 1988 members had ‘repeatedly stressed’ the need to eliminate as many ‘redundant duplicates in European collections’ as possible by systematic comparison of collections (Knüpffer, 1988a: 144). A second phase of the project was therefore to implement the database in precisely this way. The database would be used not only to identify where duplicates occurred but also to assign and track responsibility for maintaining the accessions remaining after redundant duplicates were eliminated.
The working group hoped that removing duplicate samples would be a step in the rationalisation of collection management and therefore also a step towards decentralisation. However, the identification of duplicates in the emerging database was itself ‘a time-consuming procedure requiring much knowledge about the breeding and collecting history of a particular crop’ (Knüpffer, 1988a: 150). De-duplication demanded data and new forms of data analysis. For a few individuals involved in the creation and curation of the European Barley Database, weeding out duplicates efficiently and effectively in the name of rationalisation became its own area of research. Duplicates were not a single class but comprised different types of genetic duplication, depending on their origin. ‘Identical duplication’ happened when a well-mixed sample was split in two, as happened for example in the creation of safety duplicates for off-site storage. ‘Common duplication’ referred to accessions arising from the same sample, for example when a new generation was grown out in order to renew a dwindling stock or multiply the seed to share beyond the bank. There were also ‘partial’ duplicates, ‘compound’ duplicates, and other known circumstances in which the genetic identify of samples overlapped significantly (van Hintum & Knüpffer, 1995: 128–129). (See Fig. 2).
Eliminating duplicates in the seed bank first required a knowledge of how they typically originated and their genetic relationship to the original sample. (From van Hintum & Knüpffer, 1995). Used with permission of Kluwer Academic Publishers, permission conveyed through Copyright Clearance Center, Inc.
Passport data—which, to reiterate, formed the foundation of the first generation of electronic databases of gene bank accessions—could not be used to discover many of these kinds of duplication. At the most basic level, the genetic makeup of a sample might have shifted during regeneration. The environmental conditions of grow-out, the size of the original population, mismanagement of seed lots—all these conditions and more could lead to divergence from one generation to the next. As a result, even ‘common’ duplicates (created for example by splitting a sample and sharing among two institutions) might actually become genetically distinct despite the fact that their identifying information remained exactly the same. Duplicates identified through passport data were therefore only ‘probable’ duplicates, genetically speaking, and not known duplicates (van Hintum & Knüpffer, 1995: 128). This limitation was compounded by fact that passport data were notoriously unreliable. Two scientists working on the database noted, as common occurrences, the ‘omission of (parts of) the collection number or other collection data, errors in interpretation… ty** errors, probable translation, transcription or transliteration errors or inconsistencies’ (van Hintum & Visser, 1995: 137). The records, in other words, were too messy to be trusted. Samples with the same label might in fact be genetically different, while samples with different labels might be identical.
Grow-outs and evaluations could potentially resolve the accession-identity issues plaguing the database. However, the whole promise of using the database for collection rationalisation was that it would avoid, among other things, these often-costly activities. The European Barley Database developers therefore sought to engineer around poor data, and trialled different means of identifying duplicates that took into consideration probable errors and inconsistencies. The ‘Soundex’ method of locating accessions carrying phonetically similar labels, for example ‘Closess IV’, ‘Colcess’, ‘Colcess IV’, ‘Colchicum’, ‘Colses’, and ‘Colsess’. Meanwhile the ‘Keyword in Context’ approach sussed out accessions with similar identifying information even if this hadn’t been standardized to the same database fields (see discussion in van Hintum & Knüpffer, 1995). But even where these approaches could be considered successful in that they guided database managers to probable duplicates, understanding whether identified items could be considered genetically identical (or nearly so) still required biochemical intervention (van Hintum & Visser, 1995: 143–144). There were limits, then, to the promise of streamlined de-duplication via the database.
Decentralisation nonetheless remained a key goal, and the European Barley Database was seen as the central means of achieving it. In 1997, a statement generated by the Barley Working Group outlined the imperatives for creating a ‘decentralized European Barley Collection’. It noted overall reductions in funding that negatively affected genetic resources work and emphasised that this ‘strained economic situation is further aggravated by the duplication of both efforts and germplasm’. These ‘economic constraints’ in turn required not only priority setting but also ‘the sharing of responsibilities’ and, more generally, recognition that ‘no single country in Europe can, on its own, conserve all barley genetic resources’ (Maggioni et al., 1997: 111). That same year, a new version of the European Barley Database was released, now including more than 90,000 accessions; this represented the first update since the 1987 iteration and was made possible, after efforts to secure external funding failed, by local provision of a staff person to conduct the update over a six-month period (Knüpffer et al., 2001: 50). As in the case of its predecessor, the ‘wide coverage and completeness of data’ was touted as ‘essential’ to its full use, including ‘a screening of the collections in Europe, made to identify unique samples or to locate duplicates’ (Maggioni et al., 1997: 3). Ten years on, the identified problems and solutions remained much the same. But realisation of rationalisation remained elusive.
5 Conclusions. Out of Many, One?
At a 2000 meeting of the Barley Working Group, membership of which had by that time reached more than 30 scientists from institutions across Europe and beyond, participants discussed a proposal that had been floated at higher levels of the European Cooperative Programme for Crop Genetic Resources. This was the aspiration ‘to build virtually a decentralized European Genebank’—a new formulation of what had really been the aspiration of the programme all along. The recorded discussion of this proposal among the Barley Working Group reveals members’ scepticism about its feasibility, but curiously not because of the challenges of data creation and management that limited the horizons of their own decentralisation efforts up to that point. Problems were envisioned with gene banks whose assigned accessions were not particularly useful to its core users, who would then be forced to look abroad for items of interest. Recommendations for enrolling national collections in the broader European Cooperative Initiative centred on identifying the accessions to be pooled and assigning specific accessions to the care of specific gene banks, rather than pointing out the significant investments that would be required to make this cost-cutting measure feasible (Knüpffer et al., 2001: 9–10).
Meanwhile, an expansion of the European Barley Database to link with other international collections brought the total number of accessions to more than 135,000 and made it an increasingly international, as opposed to European, enterprise. Although locating duplicates was listed as a key outcome of this further database development, the payoff of this identification was not in creating opportunities for eliminating redundancy, but simply better data sharing, ‘allow[ing] links to be established between accessions and their evaluation data accessible in the respective databases’ (Dirk & Knüpffer, 2001: 52). It is this vision of databases—as tools for sharing information, linking communities, pooling knowledge—that predominates in both celebratory and critical accounts of seed and gene banks’ database development projects. However, as I have shown here, these projects have also been driven by desire for greater economy in the expenditure of scarce resources and ironically forestalled for lack of funds. They have gained traction as political initiatives, without consistent appreciation for the technical and political challenges of realising data linkage. This brings to the fore a different narrative about the history and politics of seed and gene banks and of the data infrastructures associated with these.
Data creation, harmonisation and centralisation remain key objectives across the agricultural sciences, perhaps even more so in an era of ‘Big Data’ than in the period covered by this chapter (see, e.g., Harper et al., 2018; Arnaud et al., 2020). In the intervening years, the transformation of technical capacities has made it easier to implement forms of data linkage that could only be aspirational in the mid-to-late twentieth century. Political and economic constraints nonetheless remain a significant concern in database development, as several contributions to this volume highlight (see also Leonelli, Forthcoming). Meanwhile many crucial domains of technical skill, such as data curatorship, remain undervalued (Leonelli, 2014; Strasser, 2019; Leonelli & Tempini, 2020). If recent history points to constancy in the vision of data linkage as a solution to the imperatives of international agricultural research and development, thereby affirming contemporary calls to resolve—finally—the technical obstacles to it, this history also points to the extent to which these technical projects were and are much more than that. They have served as means of deflecting criticism, vehicles for fostering geopolitical ties, cost-cutting measures, and more. Recognising these aims, which sometimes converge but may also be in contradiction, is crucial to forging effective and equitable programmes for gene bank data management in the future.
Notes
- 1.
By the second meeting of the ECP/GR governing body in 1981, 20 countries had formally agreed to the program: Bulgaria, Cyprus, Denmark, Finland, the German Democratic Republic, Greece, Hungary, Iceland, Israel, Italy, the Netherlands, Norway, Poland, Portugal, Romania, Spain, Sweden, Switzerland, the United Kingdom, and Yugoslavia. In addition, Belgium, France, and the Federal Republic of Germany had indicated their intention to participate (ECP/GR, 1981b: 23).
- 2.
Documents prepared for the 1983 meeting of the Barley Working Group suggested that more than 17,000 barley accessions were held by the VIR in Leningrad, which (assuming the numbers were even reasonably accurate) would have made this the largest collection not only in Europe but also worldwide.
References
Aoki, K. (2008). Seed wars: Controversies and cases on plant genetic resources and intellectual property. Carolina University Press.
Arnaud, E., et al. (2020). The ontologies community of practice: A CGIAR initiative for big data in agrifood systems. Patterns, 1(7), 100105.
Barley Working Group. (1983). Report of a Working Group held at the Zentralinstitut für Genetik und Kulturpflanzenforschung der Akademie der Wissenschaften der DDR Gatersleben (AGP:IBPGR/83/68). UNDP–IBPGR.
Bonneuil, C. (2019). Seeing nature as a ‘Universal store of genes’: How biological diversity became ‘Genetic resources’. Studies in the History and Philosophy of Science Part C, 75, 1–14.
Brockway, L. (1979). Science and colonial expansion: The role of the British Royal Botanic Gardens (Reprint, 2002). Yale University Press.
Brown, A. H. D. (1989). Core collections: A practical approach to genetic resources management. Genome, 31, 818–824.
Brown, A. H. D. (1995). The core collection at the crossroads. In T. Hodgkin et al. (Eds.), Core collections of plant genetic resources (pp. 3–19). IPGRI.
Brown, A. H. D., & Spillane, C. (1999). Implementing core collections: Principles, procedures, progress, problems and promise. In R. C. Johnson & T. Hodgkin (Eds.), Core collections for today and tomorrow. IPGRI.
Chacko, X. (2019). Creative practices of care: The subjectivity, agency, and affective labor of preparing seeds for long-term banking. Culture, Agriculture, Food and Environment, 41(2), 97–106.
Curry, H. A. (2017). From working collections to the World Germplasm Project: Agricultural modernization and genetic conservation at the Rockefeller Foundation. History and Philosophy of the Life Sciences, 39, Article 5.
Curry, H. A. (2022). Endangered maize: Industrial agriculture and the crisis of extinction. University of California Press.
Curry, H. A., & Leonelli, S. (Forthcoming). Describing crops in the CGIAR era. In H. Curry & T. Lorek (Eds.), Research as development: Historical perspectives on agricultural science and international aid in the CGIAR era.
Dirk, E., & Knüpffer, H. (2001). Appendix I. The European Barley Database. In H. Knüpffer et al. (Eds.), Report of a Working Group on Barley. Sixth meeting, 3 December 2000 (pp. 50–63). IPGRI.
Drayton, R. (2000). Nature’s government: Science, Imperial Britain, and the ‘improvement’ of the world. Yale University Press.
ECP/GR [FAO/UNDP European Cooperative Program for Conservation and Exchange of Crop Genetic Resources]. (1981a). Report of the first governing board meeting, 15–18 December 1980. UNDP and FAO.
ECP/GR [FAO/UNDP European Cooperative Program for Conservation and Exchange of Crop Genetic Resources]. (1981b). Report of the second governing board meeting, 14–18 December 1981. UNDP and FAO.
ECP/GR [FAO/UNDP Cooperative Programme for Conservation and Exchange of Plant Genetic Resources]. (1982). Report of extra-ordinary meeting of the governing board, Geneva, 14–16 June 1982. UNDP and FAO.
FAO [UN Food and Agriculture Organization]. (1979). Report of the FAO/UNDP government consultation on the European Cooperative Programme for the Conservation and Exchange of Genetic Resources for Plant Breeding, Rome, 8–9 March 1979. FAO.
FAO [UN Food and Agriculture Organization]. (1980). Report of the FAO/UNDP governments consultation on the European Cooperative Programme for the Conservation and Exchange of Genetic Resources for Plant Breeding, Geneva, 17–19 December 1979. FAO.
FAO [UN Food and Agriculture Organization]. (2010). The second report on the state of the world’s plant genetic resources for food and agriculture. FAO.
Fenzi, M. (Forthcoming). Crop diversity under CGIAR’s lens. In H. Curry & T. Lorek (Eds.), Research as development: historical perspectives on agricultural science and international aid in the CGIAR era.
Fenzi, M., & Bonneuil, C. (2016). From ‘genetic resources’ to ‘ecosystems services’: A century of science and global policies for crop diversity conservation. Culture, Agriculture, Food and Environment, 38(2), 72–83.
Flitner, M. (2003). Genetic geographies: A historical comparison of agrarian modernization and eugenic thought in Germany, the Soviet Union, and the United States. Geoforum, 34(2), 175–185.
Frankel, O. H. (1984). Genetic perspectives of germplasm conservation. In W. Arber, K. Illemensee, W. J. Peacock, & P. Starlinger (Eds.), Genetic manipulation: Impact on man and society (pp. 161–170). Cambridge University Press.
Fullilove, C. (2018). Microbiology and the imperatives of capital in international agro-biodiversity preservation. Osiris, 33(1), 294–318.
Goodman, M. M. (1984). An evaluation and critique of current germplasm programs. In Report of the 1983 Plant Breeding Research Forum (pp. 195–249). Pioneer Hi-Bred International.
Hanson, J., Williams, J. T., & Freund, R. (1984). Institutes conserving crop germplasm: The IBPGR global network of genebanks. IPBGR.
Harper, L., et al. (2018). AgBioData Consortium recommendations for sustainable genomics and genetics databases for agriculture. Database, 2018, bay088.
Knüpffer, H. (1988a). The European Barley Database of the ECP/GR: An introduction. Kulturpflanze, 36, 135–162.
Knüpffer, H. (1988b). The European Barley Data Base of the European Cooperative Programme for the Conservation and Exchange of Crop Genetic Resources (ECP/GR). Plant Genetic Resources Newsletter, 75(76), 17–20.
Knüpffer, H., et al. (2001). Report of a Working Group on Barley, sixth meeting, 3 December 2000. IPGRI.
Lehmann, C. O. (1981). Collecting European land-races and development of European gene banks-historical remarks. Die Kulturpflanze, 29(1), 29–40.
Leonelli, S. (2014). Data-centric biology: A philosophical study. University of Chicago Press.
Leonelli, S. (Forthcoming). How data cross borders: Globalising plant knowledge through transnational data management and its epistemic economy. In J. Krige (Ed.), Transnational transactions: Negotiating the movement of knowledge across borders. University of Chicago Press.
Leonelli, S., & Tempini, N. (2020). Data journeys in the sciences. Springer.
Lyman, J. M. (1984). Progress and planning for germplasm conservation of major food crops. Plant Genetic Resources Newsletter, 60, 3–21.
Maggioni, L., et al. (1997). Report of a Working Group on Barley, fifth meeting, 10–12 July 1997. IPGRI.
Mooney, P. R. (1979). Seeds of the earth: A private or public resource? International Coalition for Development Action.
Mooney, P. R. (1983). The law of the seed. Development Dialogue, 1–2, 1–172.
National Research Council (NRC) [United States]. (1993). Managing global genetic resources: Agricultural crop issues and policies. The National Academies Press.
Parry, B. (2004). Trading the genome: Investigating the commodification of bio-information. Columbia University Press.
Peeters, J. P., & Williams, J. T. (1984). Towards better use of genebanks with special reference to information. Plant Genetic Resources Newsletter, 60, 22–32.
Peres, S. (2019). Seed banking as cryopower: A cryopolitical account of the work of the International Board of Plant Genetic Resources, 1973–1984. Culture, Agriculture, Food & Environment, 41(2), 76–86.
Perret, P. M. (1985). European Programme for Conservation and Exchange of Crop Genetic Resources. Plant Genetic Resources Newsletter, 61, 5–12.
Pistorius, R. (1997). Scientists, plants and politics: A history of the plant genetic resources movement. IPGRI.
Pistorius, R., & van Wijk, J. (1997). The exploitation of plant genetic information: Political strategies in crop development. CABI.
Saraiva, T. (2013). Breeding Europe: Crop diversity, gene banks, and commoners. In N. Disco & E. Kranakis (Eds.), Cosmopolitan commons: Sharing resources and risks across borders (pp. 185–212). MIT Press.
Strasser, B. (2019). Collecting experiments: Making big data biology. University of Chicago Press.
Thormann, I., Engels, J. M. M., & Halewood, M. (2019). Are the old International Board for Plant Genetic Resources (IBPGR) base collections available through the Plant Treaty’s multilateral system of access and benefit sharing? A review. Genetic Resoures and Crop Evolution, 66, 291–310.
UNDP-IBPGR [United Nations Development Programme-International Board for Plant Genetic Resources]. (1984). The ECP/GR: An introduction to the European Cooperative Programme for the Conservation and Exchange of Plant Genetic Resources. IBPGR.
US Comptroller General. (1981). Report to the Congress of the United States: The Department of Agriculture can minimize the risk of potential crop failures (CED-81-75, 10 April). US General Accounting Office.
Van Dooren, T. (2010). Banking seed: Use and value in the conservation of agricultural diversity. Science as Culture, 18(4), 373–395.
van Hintum, T. J. L., & Knüpffer, H. (1995). Duplication within and between germplasm collections. I. Identifying duplication on the basis of passport data. Genetic Resources and Crop Evolution, 42, 127–133.
van Hintum, T. J. L., & Visser, D. L. (1995). Duplication within and between germplasm collections. II. Duplication in four European barley collections. Genetic Resources and Crop Evolution, 42, 135–145.
Weise, S., Lohwasser, U., & Oppermann, M. (2020). Document or lose it – On the importance of information management for genetic resources conservation in Genebanks. Plants, 9(8), 1050. https://doi.org/10.3390/plants9081050
Williams, J. T. (1984). A decade of crop genetic resources research. In J. H. W. Holden & J. T. Williams (Eds.), Crop genetic resources: Conservation & evaluation (pp. 1–17). George Allen & Unwin.
Acknowledgements
This research was funded by the Wellcome Trust [Grant number 217968/Z/19/Z]. For the purpose of open access, the author has applied a CC BY public copyright licence to any Author Accepted Manuscript version arising from this submission.
The author is grateful to Jessica J. Lee for her help researching this article, to workshop participants and members of the ‘From Collection to Cultivation’ project team for helpful feedback on an early draft, and to Sabina Leonelli and Hugh Williamson for their editorial direction.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2023 The Author(s)
About this chapter
Cite this chapter
Curry, H.A. (2023). Data, Duplication, and Decentralisation: Gene Bank Management in the 1980s and 1990s. In: Williamson, H.F., Leonelli, S. (eds) Towards Responsible Plant Data Linkage: Data Challenges for Agricultural Research and Development. Springer, Cham. https://doi.org/10.1007/978-3-031-13276-6_9
Download citation
DOI: https://doi.org/10.1007/978-3-031-13276-6_9
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-13275-9
Online ISBN: 978-3-031-13276-6
eBook Packages: Religion and PhilosophyPhilosophy and Religion (R0)