Abstract
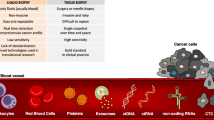
Metastatic dissemination accounts for most of the death in patients during cancer progression. There is thus an urge to identify specific biomarkers as proxies for cancer progression and assessment of treatment efficiency. Cancer is a systemic disease involving the shuttling of tumor cells and tumor secreted factors to distant organs, mostly via biofluids. During this transfer, these factors are accessible for easy sampling and therefore constitute a unique source of information witnessing the presence and the evolution of the disease. Hence, liquid biopsies offer multiple advantages, including simple and low-invasive sampling procedures, low cost, and higher compliance. Importantly, liquid biopsies are adapted to personalized medicine allowing a longitudinal follow-up to monitor treatment efficiency or resistance, and risk of relapse.
The evolution of methodologies to isolate circulating tumor cells (CTCs) and extracellular vesicles (EVs) from blood samples associated with the characterization of their membrane surface repertoire and content have been instrumental in the emergence of liquid biopsies as an easy and non-invasive alternative as opposed to classical surgery-mediated tumor biopsies.
In this chapter, we comment on CTCs and EVs carrying features with great potential as cancer biomarkers. More specifically, we focus on the adhesive and mechanical properties of CTCs as metastatic markers. We also consider the recent development of EVs isolation methods and the identification of new biomarkers. Finally, we discuss their relevance as cancer prognosis tools.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Srivastava S, Koay EJ, Borowsky AD et al (2019) Cancer overdiagnosis: a biological challenge and clinical dilemma. Nat Rev Cancer 19:349–358. https://doi.org/10.1038/S41568-019-0142-8
Siegel RL, Miller KD, Jemal A (2020) Cancer statistics, 2020. CA Cancer J Clin 70:7–30. https://doi.org/10.3322/CAAC.21590
Alix-Panabières C (2020) The future of liquid biopsy. Nature 579:S9. https://doi.org/10.1038/D41586-020-00844-5
Alix-Panabières C, Pantel K (2021) Liquid biopsy: from discovery to clinical application. Cancer Discov 11:858–873. https://doi.org/10.1158/2159-8290.CD-20-1311
Zhou E, Li Y, Wu F et al (2021) Circulating extracellular vesicles are effective biomarkers for predicting response to cancer therapy. EBioMedicine 67:103365. https://doi.org/10.1016/j.ebiom.2021.103365
Lambert AW, Pattabiraman DR, Weinberg RA (2017) Emerging biological principles of metastasis. Cell 168:670–691. https://doi.org/10.1016/j.cell.2016.11.037
Friedl P, Wolf K, Lammerding J (2011) Nuclear mechanics during cell migration. Curr Opin Cell Biol 23:55–64
Eddy RJ, Weidmann MD, Sharma VP, Condeelis JS (2017) Tumor cell invadopodia: invasive protrusions that orchestrate metastasis. Trends Cell Biol 27:595–607. https://doi.org/10.1016/j.tcb.2017.03.003
Ozimski LL, Gremmelspacher D, Aceto N (2021) A fatal affair: circulating tumor cell relationships that shape metastasis. iScience 24:103073. https://doi.org/10.1016/j.isci.2021.103073
Follain G, Herrmann D, Harlepp S et al (2020) Fluids and their mechanics in tumour transit: sha** metastasis. Nat Rev Cancer 20:107–124. https://doi.org/10.1038/s41568-019-0221-x
Obenauf AC, Massagué J (2015) Surviving at a distance: organ-specific metastasis. Trends Cancer 1:76–91. https://doi.org/10.1016/j.trecan.2015.07.009
Klein CA (2020) Cancer progression and the invisible phase of metastatic colonization. Nat Rev Cancer 20:681–694. https://doi.org/10.1038/s41568-020-00300-6
Phan TG, Croucher PI (2020) The dormant cancer cell life cycle. Nat Rev Cancer 20:398–411. https://doi.org/10.1038/s41568-020-0263-0
Dongre A, Weinberg RA (2019) New insights into the mechanisms of epithelial–mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol 20:69. https://doi.org/10.1038/s41580-018-0080-4
Pastushenko I, Blanpain C (2019) EMT transition states during tumor progression and metastasis. Trends Cell Biol 29:212–226. https://doi.org/10.1016/j.tcb.2018.12.001
Brabletz S, Schuhwerk H, Brabletz T, Stemmler MP (2021) Dynamic EMT: a multi-tool for tumor progression. EMBO J 40. https://doi.org/10.15252/embj.2021108647
Lambert AW, Weinberg RA (2021) Linking EMT programmes to normal and neoplastic epithelial stem cells. Nat Rev Cancer 21:325–338. https://doi.org/10.1038/s41568-021-00332-6
Massagué J, Ganesh K (2021) Metastasis-initiating cells and ecosystems. Cancer Discov 11:971–994. https://doi.org/10.1158/2159-8290.CD-21-0010
Fabbiano F, Corsi J, Gurrieri E et al (2020) RNA packaging into extracellular vesicles: an orchestra of RNA-binding proteins? J Extracell Vesicles. https://doi.org/10.1002/jev2.12043
van Niel G, D’Angelo G, Raposo G (2018) Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. https://doi.org/10.1038/nrm.2017.125
Skotland T, Hessvik NP, Sandvig K, Llorente A (2019) Exosomal lipid composition and the role of ether lipids and phosphoinositides in exosome biology. J Lipid Res 60:9–18. https://doi.org/10.1194/jlr.R084343
Balaj L, Lessard R, Dai L et al (2011) Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat Commun. https://doi.org/10.1038/ncomms1180
Kahlert C, Melo SA, Protopopov A et al (2014) Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J Biol Chem 289:3869–3875. https://doi.org/10.1074/jbc.C113.532267
Möller A, Lobb RJ (2020) The evolving translational potential of small extracellular vesicles in cancer. Nat Rev Cancer 20:697–709. https://doi.org/10.1038/s41568-020-00299-w
Kalluri R, LeBleu VS (2020) The biology, function, and biomedical applications of exosomes. Science 367. https://doi.org/10.1126/science.aau6977
Sheehan C, D’Souza-Schorey C (2019) Tumor-derived extracellular vesicles: molecular parcels that enable regulation of the immune response in cancer. J Cell Sci 132(20):jcs235085
Ghoroghi S, Mary B, Asokan N et al (2021) Tumor extracellular vesicles drive metastasis (it’s a long way from home). FASEB BioAdvances 3:930–943. https://doi.org/10.1096/FBA.2021-00079
Peinado H, Zhang H, Matei IR et al (2017) Pre-metastatic niches: organ-specific homes for metastases. Nat Rev Cancer 17:302–317. https://doi.org/10.1038/nrc.2017.6
Arraud N, Linares R, Tan S et al (2014) Extracellular vesicles from blood plasma: determination of their morphology, size, phenotype and concentration. J Thromb Haemost 12:614–627. https://doi.org/10.1111/JTH.12554
Sódar BW, Kittel Á, Pálóczi K et al (2016) Low-density lipoprotein mimics blood plasma-derived exosomes and microvesicles during isolation and detection. Sci Rep 6:1–12. https://doi.org/10.1038/srep24316
Klein CA (2013) Selection and adaptation during metastatic cancer progression. Nature 501:365–372. https://doi.org/10.1038/nature12628
Naxerova K, Jain RK (2015) Using tumour phylogenetics to identify the roots of metastasis in humans. Nat Rev Clin Oncol 12:258–272. https://doi.org/10.1038/nrclinonc.2014.238
Hu Z, Curtis C (2020) Looking backward in time to define the chronology of metastasis. Nat Commun 11:3213. https://doi.org/10.1038/s41467-020-16995-y
Steeg PS (2016) Targeting metastasis. Nat Rev Cancer 16:201–218. https://doi.org/10.1038/nrc.2016.25
Esposito M, Ganesan S, Kang Y (2021) Emerging strategies for treating metastasis. Nat Cancer 2:258–270. https://doi.org/10.1038/s43018-021-00181-0
Ganesh K, Massagué J (2021) Targeting metastatic cancer. Nat Med 27:34–44. https://doi.org/10.1038/s41591-020-01195-4
Bardelli A, Pantel K (2017) Liquid biopsies, what we do not know (yet). Cancer Cell 31:172–179. https://doi.org/10.1016/j.ccell.2017.01.002
Keller L, Pantel K (2019) Unravelling tumour heterogeneity by single-cell profiling of circulating tumour cells. Nat Rev Cancer 19:553–567. https://doi.org/10.1038/s41568-019-0180-2
Stahel RA, Gilks WR, Lehmann H-P, Schenker T (1994) Third international workshop on lung tumor and differentiation antigens: overview of the results of the central data analysis. Int J Cancer 57:6–26. https://doi.org/10.1002/ijc.2910570704
Cristofanilli M, Budd GT, Ellis MJ et al (2004) Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med 351:781–791. https://doi.org/10.1056/NEJMoa040766
Allard WJ, Matera J, Miller MC et al (2004) Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res 10:6897–6904. https://doi.org/10.1158/1078-0432.CCR-04-0378
Kang Y-T, Hadlock T, Lo T-W et al (2020) Dual-isolation and profiling of circulating tumor cells and cancer exosomes from blood samples with melanoma using immunoaffinity-based microfluidic interfaces. Adv Sci 7:2001581. https://doi.org/10.1002/advs.202001581
Mostert B, Kraan J, Bolt-de Vries J et al (2011) Detection of circulating tumor cells in breast cancer may improve through enrichment with anti-CD146. Breast Cancer Res Treat 127:33–41. https://doi.org/10.1007/s10549-010-0879-y
Onstenk W, Kraan J, Mostert B et al (2015) Improved circulating tumor cell detection by a combined EpCAM and MCAM CellSearch enrichment approach in patients with breast cancer undergoing neoadjuvant chemotherapy. Mol Cancer Ther 14:821–827. https://doi.org/10.1158/1535-7163.MCT-14-0653
Nagrath S, Sequist LV, Maheswaran S et al (2007) Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature 450:1235–1239. https://doi.org/10.1038/nature06385
Stott SL, Hsu C-H, Tsukrov DI et al (2010) Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc Natl Acad Sci 107:18392–18397. https://doi.org/10.1073/pnas.1012539107
Diener J, Sommer L (2021) Reemergence of neural crest stem cell-like states in melanoma during disease progression and treatment. Stem Cells Transl Med 10:522–533. https://doi.org/10.1002/sctm.20-0351
Mani SA, Guo W, Liao M-J et al (2008) The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 133:704–715. https://doi.org/10.1016/j.cell.2008.03.027
Yu M, Bardia A, Wittner BS et al (2013) Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 339:580–584. https://doi.org/10.1126/science.1228522
Sieuwerts AM, Kraan J, Bolt J et al (2009) Anti-epithelial cell adhesion molecule antibodies and the detection of circulating normal-like breast tumor cells. J Natl Cancer Inst 101:61–66. https://doi.org/10.1093/jnci/djn419
Chaffer CL, Brueckmann I, Scheel C et al (2011) Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc Natl Acad Sci 108:7950–7955. https://doi.org/10.1073/pnas.1102454108
Zhang L, Ridgway LD, Wetzel MD et al (2013) The identification and characterization of breast cancer CTCs competent for brain metastasis. Sci Transl Med 5:180ra48. https://doi.org/10.1126/scitranslmed.3005109
Alix-Panabières C, Vendrell J-P, Slijper M et al (2009) Full-length cytokeratin-19 is released by human tumor cells: a potential role in metastatic progression of breast cancer. Breast Cancer Res 11:1–10. https://doi.org/10.1186/bcr2326
Denève E, Riethdorf S, Ramos J et al (2013) Capture of viable circulating tumor cells in the liver of colorectal cancer patients. Clin Chem 59:1384–1392. https://doi.org/10.1373/clinchem.2013.202846
Donati G, Watt FM (2015) Stem cell heterogeneity and plasticity in epithelia. Cell Stem Cell 16:465–476. https://doi.org/10.1016/j.stem.2015.04.014
Jahchan NS, Lim JS, Bola B et al (2016) Identification and targeting of long-term tumor-propagating cells in small cell lung cancer. Cell Rep 16:644–656. https://doi.org/10.1016/j.celrep.2016.06.021
Al-Hajj M, Wicha MS, Benito-Hernandez A et al (2003) Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci 100:3983–3988. https://doi.org/10.1073/pnas.0530291100
Chaffer CL, Marjanovic ND, Lee T et al (2013) Poised chromatin at the ZEB1 promoter enables breast cancer cell plasticity and enhances tumorigenicity. Cell 154:61–74. https://doi.org/10.1016/j.cell.2013.06.005
Taftaf R, Liu X, Singh S et al (2021) ICAM1 initiates CTC cluster formation and trans-endothelial migration in lung metastasis of breast cancer. Nat Commun 12:4867. https://doi.org/10.1038/s41467-021-25189-z
Ward Y, Lake R, Faraji F et al (2018) Platelets promote metastasis via binding tumor CD97 leading to bidirectional signaling that coordinates transendothelial migration. Cell Rep 23:808–822. https://doi.org/10.1016/j.celrep.2018.03.092
Eibl RH (2012) Single-molecule studies of integrins by AFM-based force spectroscopy on living cells. In: Scanning probe microscopy in nanoscience and nanotechnology, vol 3. Springer, pp 137–169
Oatley M, Bölükbası ÖV, Svensson V et al (2020) Single-cell transcriptomics identifies CD44 as a marker and regulator of endothelial to haematopoietic transition. Nat Commun 11:1–18. https://doi.org/10.1038/s41467-019-14171-5
Wang T, Ward Y, Tian L et al (2005) CD97, an adhesion receptor on inflammatory cells, stimulates angiogenesis through binding integrin counterreceptors on endothelial cells. Blood 105:2836–2844. https://doi.org/10.1182/blood-2004-07-2878
Humphries JD, Byron A, Humphries MJ (2006) Integrin ligands at a glance. J Cell Sci 119:3901–3903. https://doi.org/10.1242/jcs.03098
Vishnoi M, Peddibhotla S, Yin W et al (2015) The isolation and characterization of CTC subsets related to breast cancer dormancy. Sci Rep 5:17533. https://doi.org/10.1038/srep17533
Boral D, Vishnoi M, Liu HN et al (2017) Molecular characterization of breast cancer CTCs associated with brain metastasis. Nat Commun 8:196. https://doi.org/10.1038/s41467-017-00196-1
Osmani N, Labouesse M (2015) Remodeling of keratin-coupled cell adhesion complexes. Curr Opin Cell Biol 32:30–38. https://doi.org/10.1016/j.ceb.2014.10.004
Meyer MJ, Fleming JM, Lin AF et al (2010) CD44posCD49fhiCD133/2hi defines xenograft-initiating cells in estrogen receptor–negative breast cancer. Cancer Res 70:4624–4633. https://doi.org/10.1158/0008-5472.CAN-09-3619
Bierie B, Pierce SE, Kroeger C et al (2017) Integrin-β4 identifies cancer stem cell-enriched populations of partially mesenchymal carcinoma cells. Proc Natl Acad Sci 114:E2337–E2346. https://doi.org/10.1073/pnas.1618298114
Sharifi M, Zarrin B, Najafi MB et al (2021) Integrin α6 β4 on circulating tumor cells of metastatic breast cancer patients. Adv Biomed Res 10:16. https://doi.org/10.4103/abr.abr_76_21
Kröger C, Afeyan A, Mraz J et al (2019) Acquisition of a hybrid E/M state is essential for tumorigenicity of basal breast cancer cells. Proc Natl Acad Sci 116:7353–7362. https://doi.org/10.1073/pnas.1812876116
Ganesh K, Basnet H, Kaygusuz Y et al (2020) L1CAM defines the regenerative origin of metastasis-initiating cells in colorectal cancer. Nat Cancer 1:28–45. https://doi.org/10.1038/s43018-019-0006-x
Padmanaban V, Krol I, Suhail Y et al (2019) E-cadherin is required for metastasis in multiple models of breast cancer. Nature 573:439–444. https://doi.org/10.1038/s41586-019-1526-3
Na T-Y, Schecterson L, Mendonsa AM, Gumbiner BM (2020) The functional activity of E-cadherin controls tumor cell metastasis at multiple steps. Proc Natl Acad Sci 117:5931–5937. https://doi.org/10.1073/pnas.1918167117
Hapach LA, Carey SP, Schwager SC et al (2021) Phenotypic heterogeneity and metastasis of breast cancer cells. Cancer Res 81:3649–3663. https://doi.org/10.1158/0008-5472.CAN-20-1799
Fang C, Fan C, Wang C et al (2016) CD133 + CD54 + CD44 + circulating tumor cells as a biomarker of treatment selection and liver metastasis in patients with colorectal cancer. Oncotarget 7:77389–77403. https://doi.org/10.18632/oncotarget.12675
Battula VL, Shi Y, Evans KW et al (2012) Ganglioside GD2 identifies breast cancer stem cells and promotes tumorigenesis. J Clin Invest 122:2066–2078. https://doi.org/10.1172/JCI59735
Liang Y-J, Ding Y, Levery SB et al (2013) Differential expression profiles of glycosphingolipids in human breast cancer stem cells vs. cancer non-stem cells. Proc Natl Acad Sci 110:4968–4973. https://doi.org/10.1073/pnas.1302825110
Chiu CG, Nakamura Y, Chong KK et al (2014) Genome-wide characterization of circulating tumor cells identifies novel prognostic genomic alterations in systemic melanoma metastasis. Clin Chem 60:873–885. https://doi.org/10.1373/clinchem.2013.213611
Fasanya HO, Dopico PJ, Yeager Z et al (2021) Using a combination of gangliosides and cell surface vimentin as surface biomarkers for isolating osteosarcoma cells in microfluidic devices. J Bone Oncol 28:100357. https://doi.org/10.1016/j.jbo.2021.100357
Satelli A, Mitra A, Brownlee Z et al (2015) Epithelial–mesenchymal transitioned circulating tumor cells capture for detecting tumor progression. Clin Cancer Res 21:899–906. https://doi.org/10.1158/1078-0432.CCR-14-0894
Satelli A, Batth I, Brownlee Z et al (2017) EMT circulating tumor cells detected by cell-surface vimentin are associated with prostate cancer progression. Oncotarget 8:49329–49337. https://doi.org/10.18632/oncotarget.17632
**e X, Wang L, Wang X et al (2021) Evaluation of cell surface vimentin positive circulating tumor cells as a diagnostic biomarker for lung cancer. Front Oncol 11:1712. https://doi.org/10.3389/fonc.2021.672687
Ginestier C, Hur MH, Charafe-Jauffret E et al (2007) ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell 1:555–567. https://doi.org/10.1016/j.stem.2007.08.014
Papadaki MA, Stoupis G, Theodoropoulos PA et al (2019) Circulating tumor cells with stemness and epithelial-to-mesenchymal transition features are chemoresistant and predictive of poor outcome in metastatic breast cancer. Mol Cancer Ther 18:437–447. https://doi.org/10.1158/1535-7163.MCT-18-0584
Leng Z, Yang Z, Li L et al (2017) A reliable method for the sorting and identification of ALDHhigh cancer stem cells by flow cytometry. Exp Ther Med 14:2801–2808. https://doi.org/10.3892/etm.2017.4846
Northcott JM, Dean IS, Mouw JK, Weaver VM (2018) Feeling stress: the mechanics of cancer progression and aggression. Front Cell Dev Biol 6. https://doi.org/10.3389/fcell.2018.00017
Levayer R (2020) Solid stress, competition for space and cancer: the opposing roles of mechanical cell competition in tumour initiation and growth. Semin Cancer Biol 63:69–80. https://doi.org/10.1016/j.semcancer.2019.05.004
Stylianopoulos T, Munn LL, Jain RK (2018) Reengineering the physical microenvironment of tumors to improve drug delivery and efficacy: from mathematical modeling to bench to bedside. Trends Cancer 4:292–319. https://doi.org/10.1016/j.trecan.2018.02.005
Wei SC, Yang J (2016) Forcing through tumor metastasis: the interplay between tissue rigidity and epithelial–mesenchymal transition. Trends Cell Biol 26:111–120. https://doi.org/10.1016/j.tcb.2015.09.009
Kai F, Laklai H, Weaver VM (2016) Force matters: biomechanical regulation of cell invasion and migration in disease. Trends Cell Biol 26:486–497. https://doi.org/10.1016/j.tcb.2016.03.007
Gensbittel V, Kräter M, Harlepp S et al (2021) Mechanical adaptability of tumor cells in metastasis. Dev Cell 56:164–179. https://doi.org/10.1016/j.devcel.2020.10.011
Wirtz D, Konstantopoulos K, Searson PC (2011) The physics of cancer: the role of physical interactions and mechanical forces in metastasis. Nat Rev Cancer 11:512–522. https://doi.org/10.1038/nrc3080
Ozkumur E, Shah AM, Ciciliano JC et al (2013) Inertial focusing for tumor antigen–dependent and –independent sorting of rare circulating tumor cells. Sci Transl Med 5:179ra47. https://doi.org/10.1126/scitranslmed.3005616
Alibert C, Goud B, Manneville J-B (2017) Are cancer cells really softer than normal cells? Biol Cell 109:167–189. https://doi.org/10.1111/boc.201600078
Plodinec M, Loparic M, Monnier CA et al (2012) The nanomechanical signature of breast cancer. Nat Nanotechnol 7:757–765. https://doi.org/10.1038/nnano.2012.167
Vona G, Sabile A, Louha M et al (2000) Isolation by size of epithelial tumor cells: a new method for the immunomorphological and molecular characterization of circulating tumor cells. Am J Pathol 156:57–63. https://doi.org/10.1016/S0002-9440(10)64706-2
Toner M, Irimia D (2005) Blood-on-a-chip. Annu Rev Biomed Eng 7:77–103. https://doi.org/10.1146/annurev.bioeng.7.011205.135108
Mohamed H, McCurdy LD, Szarowski DH et al (2004) Development of a rare cell fractionation device: application for cancer detection. IEEE Trans Nanobiosci 3:251–256. https://doi.org/10.1109/TNB.2004.837903
Xu L, Mao X, Imrali A et al (2015) Optimization and evaluation of a novel size based circulating tumor cell isolation system. PLoS One 10:e0138032. https://doi.org/10.1371/journal.pone.0138032
Hou HW, Warkiani ME, Khoo BL et al (2013) Isolation and retrieval of circulating tumor cells using centrifugal forces. Sci Rep 3:1259. https://doi.org/10.1038/srep01259
Miller MC, Robinson PS, Wagner C, O’Shannessy DJ (2018) The Parsortix™ cell separation system—a versatile liquid biopsy platform. Cytometry A 93:1234–1239. https://doi.org/10.1002/cyto.a.23571
Lee Y, Guan G, Bhagat AA (2018) ClearCell® FX, a label-free microfluidics technology for enrichment of viable circulating tumor cells. Cytometry A 93:1251–1254. https://doi.org/10.1002/cyto.a.23507
Mishra A, Dubash TD, Edd JF et al (2020) Ultrahigh-throughput magnetic sorting of large blood volumes for epitope-agnostic isolation of circulating tumor cells. Proc Natl Acad Sci 117:16839–16847. https://doi.org/10.1073/pnas.2006388117
Kim T-H, Lim M, Park J et al (2017) FAST: size-selective, clog-free isolation of rare cancer cells from whole blood at a liquid–liquid interface. Anal Chem 89:1155–1162. https://doi.org/10.1021/acs.analchem.6b03534
Aceto N, Bardia A, Miyamoto DT et al (2014) Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell 158:1110–1122. https://doi.org/10.1016/j.cell.2014.07.013
Gkountela S, Castro-Giner F, Szczerba BM et al (2019) Circulating tumor cell clustering shapes DNA methylation to enable metastasis seeding. Cell 176:98–112.e14. https://doi.org/10.1016/j.cell.2018.11.046
Szczerba BM, Castro-Giner F, Vetter M et al (2019) Neutrophils escort circulating tumour cells to enable cell cycle progression. Nature 566:553. https://doi.org/10.1038/s41586-019-0915-y
Sarioglu AF, Aceto N, Kojic N et al (2015) A microfluidic device for label-free, physical capture of circulating tumor cell clusters. Nat Methods 12:685–691. https://doi.org/10.1038/nmeth.3404
Au SH, Storey BD, Moore JC et al (2016) Clusters of circulating tumor cells traverse capillary-sized vessels. Proc Natl Acad Sci 113:4947–4952. https://doi.org/10.1073/pnas.1524448113
Jiang X, Wong KHK, Khankhel AH et al (2017) Microfluidic isolation of platelet-covered circulating tumor cells. Lab Chip 17:3498–3503. https://doi.org/10.1039/C7LC00654C
Guck J, Schinkinger S, Lincoln B et al (2005) Optical deformability as an inherent cell marker for testing malignant transformation and metastatic competence. Biophys J 88:3689–3698. https://doi.org/10.1529/biophysj.104.045476
Lincoln B, Schinkinger S, Travis K et al (2007) Reconfigurable microfluidic integration of a dual-beam laser trap with biomedical applications. Biomed Microdevices 9:703–710. https://doi.org/10.1007/s10544-007-9079-x
Nel I, Morawetz EW, Tschodu D et al (2021) The mechanical fingerprint of circulating tumor cells (CTCs) in breast cancer patients. Cancers 13:1119. https://doi.org/10.3390/cancers13051119
Gossett DR, Tse HTK, Lee SA et al (2012) Hydrodynamic stretching of single cells for large population mechanical phenoty**. Proc Natl Acad Sci 109:7630–7635. https://doi.org/10.1073/pnas.1200107109
Adamo A, Sharei A, Adamo L et al (2012) Microfluidics-based assessment of cell deformability. Anal Chem 84:6438–6443. https://doi.org/10.1021/ac300264v
Byun S, Son S, Amodei D et al (2013) Characterizing deformability and surface friction of cancer cells. Proc Natl Acad Sci 110:7580–7585. https://doi.org/10.1073/pnas.1218806110
Lange JR, Steinwachs J, Kolb T et al (2015) Microconstriction arrays for high-throughput quantitative measurements of cell mechanical properties. Biophys J 109:26–34. https://doi.org/10.1016/j.bpj.2015.05.029
Otto O, Rosendahl P, Mietke A et al (2015) Real-time deformability cytometry: on-the-fly cell mechanical phenoty**. Nat Methods 12:199–202. https://doi.org/10.1038/nmeth.3281
Urbanska M, Muñoz HE, Shaw Bagnall J et al (2020) A comparison of microfluidic methods for high-throughput cell deformability measurements. Nat Methods 17:587–593. https://doi.org/10.1038/s41592-020-0818-8
Nyberg KD, Hu KH, Kleinman SH et al (2017) Quantitative deformability cytometry: rapid, calibrated measurements of cell mechanical properties. Biophys J 113:1574–1584. https://doi.org/10.1016/j.bpj.2017.06.073
Bagnall JS, Byun S, Begum S et al (2015) Deformability of tumor cells versus blood cells. Sci Rep 5:18542. https://doi.org/10.1038/srep18542
Holenstein CN, Horvath A, Schär B et al (2019) The relationship between metastatic potential and in vitro mechanical properties of osteosarcoma cells. Mol Biol Cell 30:887–898. https://doi.org/10.1091/mbc.E18-08-0545
Rosendahl P, Plak K, Jacobi A et al (2018) Real-time fluorescence and deformability cytometry. Nat Methods 15:355–358. https://doi.org/10.1038/nmeth.4639
Toepfner N, Herold C, Otto O et al (2018) Detection of human disease conditions by single-cell morpho-rheological phenoty** of blood. Elife 7:e29213. https://doi.org/10.7554/eLife.29213
Hakim M, Khorasheh F, Alemzadeh I, Vossoughi M (2021) A new insight to deformability correlation of circulating tumor cells with metastatic behavior by application of a new deformability-based microfluidic chip. Anal Chim Acta 1186:339115. https://doi.org/10.1016/j.aca.2021.339115
Ribeiro-Samy S, Oliveira MI, Pereira-Veiga T et al (2019) Fast and efficient microfluidic cell filter for isolation of circulating tumor cells from unprocessed whole blood of colorectal cancer patients. Sci Rep 9:8032. https://doi.org/10.1038/s41598-019-44401-1
Lopes C, Piairo P, Chícharo A et al (2021) HER2 expression in circulating tumour cells isolated from metastatic breast cancer patients using a size-based microfluidic device. Cancers 13:4446. https://doi.org/10.3390/cancers13174446
Harouaka RA, Zhou M-D, Yeh Y-T et al (2014) Flexible micro spring array device for high-throughput enrichment of viable circulating tumor cells. Clin Chem 60:323–333. https://doi.org/10.1373/clinchem.2013.206805
Ding X, Li P, Lin S-CS et al (2013) Surface acoustic wave microfluidics. Lab Chip 13:3626–3649. https://doi.org/10.1039/C3LC50361E
Li P, Mao Z, Peng Z et al (2015) Acoustic separation of circulating tumor cells. Proc Natl Acad Sci 112:4970–4975. https://doi.org/10.1073/pnas.1504484112
Bankó P, Lee SY, Nagygyörgy V et al (2019) Technologies for circulating tumor cell separation from whole blood. J Hematol Oncol 12:48. https://doi.org/10.1186/s13045-019-0735-4
Lei KF (2020) A review on microdevices for isolating circulating tumor cells. Micromachines 11:531. https://doi.org/10.3390/mi11050531
Belotti Y, Lim CT (2021) Microfluidics for liquid biopsies: recent advances, current challenges, and future directions. Anal Chem 93:4727–4738. https://doi.org/10.1021/acs.analchem.1c00410
Nanou A, Miller MC, Zeune LL et al (2020) Tumour-derived extracellular vesicles in blood of metastatic cancer patients associate with overall survival. Br J Cancer 122:801–811. https://doi.org/10.1038/s41416-019-0726-9
Nanou A, Coumans FAW, van Dalum G et al (2018) Circulating tumor cells, tumor-derived extracellular vesicles and plasma cytokeratins in castration-resistant prostate cancer patients. Oncotarget 9:19283–19293. https://doi.org/10.18632/ONCOTARGET.25019
Gurunathan S, Kang M-H, Jeyaraj M et al (2019) Review of the isolation, characterization, biological function, and multifarious therapeutic approaches of exosomes. Cell 8:307. https://doi.org/10.3390/CELLS8040307
Van Deun J, Mestdagh P, Sormunen R et al (2014) The impact of disparate isolation methods for extracellular vesicles on downstream RNA profiling. J Extracell Vesicles 3:1–14. https://doi.org/10.3402/jev.v3.24858
Veerman RE, Teeuwen L, Czarnewski P et al (2021) Molecular evaluation of five different isolation methods for extracellular vesicles reveals different clinical applicability and subcellular origin. J Extracell Vesicles 10. https://doi.org/10.1002/JEV2.12128
Karimi N, Cvjetkovic A, Jang SC et al (2018) Detailed analysis of the plasma extracellular vesicle proteome after separation from lipoproteins. Cell Mol Life Sci. https://doi.org/10.1007/s00018-018-2773-4
Simonsen JB (2017) What are we looking at? Extracellular vesicles, lipoproteins, or both? Circ Res 121:920–922. https://doi.org/10.1161/CIRCRESAHA.117.311767
Johnsen KB, Gudbergsson JM, Andresen TL, Simonsen JB (2019) What is the blood concentration of extracellular vesicles? Implications for the use of extracellular vesicles as blood-borne biomarkers of cancer. Biochim Biophys Acta Rev Cancer 1871:109–116. https://doi.org/10.1016/j.bbcan.2018.11.006
Nordin JZ, Lee Y, Vader P et al (2015) Ultrafiltration with size-exclusion liquid chromatography for high yield isolation of extracellular vesicles preserving intact biophysical and functional properties. Nanomed Nanotechnol Biol Med 11:879–883. https://doi.org/10.1016/J.NANO.2015.01.003
Zhang X, Borg EGF, Liaci AM et al (2020) A novel three step protocol to isolate extracellular vesicles from plasma or cell culture medium with both high yield and purity. J Extracell Vesicles 9. https://doi.org/10.1080/20013078.2020.1791450
Chen Y, Zhu Q, Cheng L et al (2021) Exosome detection via the ultrafast-isolation system: EXODUS. Nat Methods 18:212–218. https://doi.org/10.1038/S41592-020-01034-X
Xu R, Greening DW, Zhu HJ et al (2016) Extracellular vesicle isolation and characterization: toward clinical application. J Clin Invest 126:1152–1162. https://doi.org/10.1172/JCI81129
Jeppesen DK, Fenix AM, Franklin JL et al (2019) Reassessment of exosome composition. Cell 177:428–445.e18. https://doi.org/10.1016/j.cell.2019.02.029
Kugeratski FG, Hodge K, Lilla S et al (2021) Quantitative proteomics identifies the core proteome of exosomes with syntenin-1 as the highest abundant protein and a putative universal biomarker. Springer
Hoshino A, Kim HS, Bojmar L et al (2020) Extracellular vesicle and particle biomarkers define multiple human cancers. Cell 182:1044–1061.e18. https://doi.org/10.1016/j.cell.2020.07.009
Liang Y, Lehrich BM, Zheng S, Lu M (2021) Emerging methods in biomarker identification for extracellular vesicle-based liquid biopsy. J Extracell Vesicles:10. https://doi.org/10.1002/jev2.12090
Welsh JA, Van Der Pol E, Arkesteijn GJA et al (2020) MIFlowCyt-EV: a framework for standardized reporting of extracellular vesicle flow cytometry experiments. J Extracell Vesicles 9. https://doi.org/10.1080/20013078.2020.1713526
Campos-Silva C, Suárez H, Jara-Acevedo R et al (2019) High sensitivity detection of extracellular vesicles immune-captured from urine by conventional flow cytometry. Sci Rep 9:1–12. https://doi.org/10.1038/s41598-019-38516-8
Rodrigues M, Richards N, Ning B et al (2019) Rapid lipid-based approach for normalization of quantum-dot-detected biomarker expression on extracellular vesicles in complex biological samples. Nano Lett 19:7623–7631
Wei P, Wu F, Kang B et al (2020) Plasma extracellular vesicles detected by single molecule array technology as a liquid biopsy for colorectal cancer. J Extracell Vesicles 9:1809765. https://doi.org/10.1080/20013078.2020.1809765
Yoshioka Y, Kosaka N, Konishi Y et al (2014) Ultra-sensitive liquid biopsy of circulating extracellular vesicles using ExoScreen. Nat Commun 5:3591. https://doi.org/10.1038/ncomms4591
He D, Ho SL, Chan HN et al (2019) Molecular-recognition-based DNA nanodevices for enhancing the direct visualization and quantification of single vesicles of tumor exosomes in plasma microsamples. Anal Chem 91:2768–2775. https://doi.org/10.1021/ACS.ANALCHEM.8B04509/SUPPL_FILE/AC8B04509_SI_001.PDF
Zhang J, Shi J, Zhang H et al (2020) Localized fluorescent imaging of multiple proteins on individual extracellular vesicles using rolling circle amplification for cancer diagnosis. J Extracell Vesicles 10:e12025. https://doi.org/10.1002/JEV2.12025
Mathew DG, Beekman P, Lemay SG et al (2020) Electrochemical detection of tumor-derived extracellular vesicles on nanointerdigitated electrodes. Nano Lett 20:820–828. https://doi.org/10.1021/ACS.NANOLETT.9B02741
Huang R, He L, **a Y et al (2019) A sensitive aptasensor based on a hemin/G-quadruplex-assisted signal amplification strategy for electrochemical detection of gastric cancer exosomes. Small Weinh Bergstr Ger:15. https://doi.org/10.1002/SMLL.201900735
Zhang W, Jiang L, Diefenbach RJ et al (2020) Enabling sensitive phenotypic profiling of cancer-derived small extracellular vesicles using surface-enhanced Raman spectroscopy nanotags. ACS Sens 5:764–771. https://doi.org/10.1021/ACSSENSORS.9B02377
Wang J, Wuethrich A, Sina AAI et al (2020) Tracking extracellular vesicle phenotypic changes enables treatment monitoring in melanoma. Sci Adv 6. https://doi.org/10.1126/SCIADV.AAX3223
Shao H, Chung J, Lee K et al (2015) Chip-based analysis of exosomal mRNA mediating drug resistance in glioblastoma. Nat Commun 6:1–9. https://doi.org/10.1038/ncomms7999
Cappello F, Logozzi M, Campanella C et al (2017) Exosome levels in human body fluids: a tumor marker by themselves? Eur J Pharm Sci 96:93–98. https://doi.org/10.1016/j.ejps.2016.09.010
García-Silva S, Benito-Martín A, Sánchez-Redondo S et al (2019) Use of extracellular vesicles from lymphatic drainage as surrogate markers of melanoma progression and BRAF V600E mutation. J Exp Med. https://doi.org/10.1084/jem.20181522
Osti D, Del Bene M, Rappa G et al (2019) Clinical significance of extracellular vesicles in plasma from glioblastoma patients. https://doi.org/10.1158/1078-0432.CCR-18-1941
Peinado H, Alečković M, Lavotshkin S et al (2012) Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med 18:883–891. https://doi.org/10.1038/nm.2753
Sabbagh Q, André-Grégoire G, Alves-Nicolau C et al (2021) The von Willebrand factor stamps plasmatic extracellular vesicles from glioblastoma patients. Sci Rep 11:1–11. https://doi.org/10.1038/s41598-021-02254-7
Venturella M, Criscuoli M, Carraro F et al (2021) Interplay between hypoxia and extracellular vesicles in cancer and inflammation. Biology 10. https://doi.org/10.3390/BIOLOGY10070606
Keklikoglou I, Cianciaruso C, Güç E et al (2019) Chemotherapy elicits pro-metastatic extracellular vesicles in breast cancer models. Nat Cell Biol 21:190–202. https://doi.org/10.1038/s41556-018-0256-3
Federici C, Petrucci F, Caimi S et al (2014) Exosome release and low pH belong to a framework of resistance of human melanoma cells to cisplatin. PLoS One 9. https://doi.org/10.1371/journal.pone.0088193
Mutschelknaus L, Peters C, Winkler K et al (2016) Exosomes derived from squamous head and neck cancer promote cell survival after ionizing radiation. PLoS One:11. https://doi.org/10.1371/JOURNAL.PONE.0152213
Zaborowski MP, Lee K, Na YJ et al (2019) Methods for systematic identification of membrane proteins for specific capture of cancer-derived extracellular vesicles. Cell Rep 27:255–268.e6. https://doi.org/10.1016/j.celrep.2019.03.003
Whitham M, Parker BL, Friedrichsen M et al (2018) Extracellular vesicles provide a means for tissue crosstalk during exercise. Cell Metab 27:237–251.e4. https://doi.org/10.1016/j.cmet.2017.12.001
Eitan E, Green J, Bodogai M et al (2017) Age-related changes in plasma extracellular vesicle characteristics and internalization by leukocytes. Sci Rep 7:1342. https://doi.org/10.1038/s41598-017-01386-z
Newman LA, Fahmy A, Sorich MJ et al (2021) Importance of between and within subject variability in extracellular vesicle abundance and cargo when performing biomarker analyses. Cell 10:1–18. https://doi.org/10.3390/CELLS10030485
Laurenzana I, Trino S, Lamorte D et al (2021) Analysis of amount, size, protein phenotype and molecular content of circulating extracellular vesicles identifies new biomarkers in multiple myeloma. Int J Nanomed 16:3141–3160. https://doi.org/10.2147/IJN.S303391
Melo SA, Luecke LB, Kahlert C et al (2015) Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 523:177–182. https://doi.org/10.1038/nature14581
Keup C, Mach P, Aktas B et al (2018) RNA profiles of circulating tumor cells and extracellular vesicles for therapy stratification of metastatic breast cancer patients. https://doi.org/10.1373/clinchem.2017.283531
Lai X, Wang M, McElyea SD et al (2017) A microRNA signature in circulating exosomes is superior to exosomal glypican-1 levels for diagnosing pancreatic cancer. Cancer Lett 393:86–93. https://doi.org/10.1016/J.CANLET.2017.02.019
Lucien F, Lac V, Billadeau DD et al (2019) Glypican-1 and glycoprotein 2 bearing extracellular vesicles do not discern pancreatic cancer from benign pancreatic diseases. Oncotarget 10:1045–1055. https://doi.org/10.18632/ONCOTARGET.26620
Hu J, Sheng Y, Kwak KJ et al (2017) A signal-amplifiable biochip quantifies extracellular vesicle-associated RNAs for early cancer detection. Nat Commun 8. https://doi.org/10.1038/S41467-017-01942-1
Li J, Chen Y, Guo X et al (2017) GPC1 exosome and its regulatory miRNAs are specific markers for the detection and target therapy of colorectal cancer. J Cell Mol Med 21:838–847. https://doi.org/10.1111/JCMM.12941
**ao D, Dong Z, Zhen L et al (2020) Combined exosomal GPC1, CD82, and serum CA19-9 as multiplex targets: a specific, sensitive, and reproducible detection panel for the diagnosis of pancreatic cancer. Mol Cancer Res 18:1300–1310. https://doi.org/10.1158/1541-7786.MCR-19-0588
Buscail E, Chauvet A, Quincy P et al (2019) CD63-GPC1-positive exosomes coupled with CA19-9 offer good diagnostic potential for resectable pancreatic ductal adenocarcinoma. Transl Oncol 12:1395–1403. https://doi.org/10.1016/J.TRANON.2019.07.009
Poggio M, Hu T, Pai CC et al (2019) Suppression of exosomal PD-L1 induces systemic anti-tumor immunity and memory. Cell 177:414–427.e13. https://doi.org/10.1016/j.cell.2019.02.016
Chen G, Huang AC, Zhang W et al (2018) Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature 560:382–386
Cordonnier M, Nardin C, Chanteloup G et al (2020) Tracking the evolution of circulating exosomal-PD-L1 to monitor melanoma patients. J Extracell Vesicles 9:1–11. https://doi.org/10.1080/20013078.2019.1710899
Zhou B, Xu K, Zheng X et al (2020) Application of exosomes as liquid biopsy in clinical diagnosis. Signal Transduct Target Ther 5. https://doi.org/10.1038/s41392-020-00258-9
Hu T, Wolfram J, Srivastava S (2021) Extracellular vesicles in cancer detection: hopes and hypes. Trends Cancer 7:122–133. https://doi.org/10.1016/j.trecan.2020.09.003
Costa-Silva B, Aiello NM, Ocean AJ et al (2015) Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat Cell Biol:1–7. https://doi.org/10.1038/ncb3169
Choi ES, Al Faruque H, Kim JH et al (2021) CD5L as an extracellular vesicle-derived biomarker for liquid biopsy of lung cancer. Diagnostics (Basel) 11. https://doi.org/10.3390/DIAGNOSTICS11040620
Moon PG, Lee JE, Cho YE et al (2016) Identification of developmental endothelial Locus-1 on circulating extracellular vesicles as a novel biomarker for early breast cancer detection. Clin Cancer Res Off J Am Assoc Cancer Res 22:1757–1766. https://doi.org/10.1158/1078-0432.CCR-15-0654
Moon PG, Lee JE, Cho YE et al (2016) Fibronectin on circulating extracellular vesicles as a liquid biopsy to detect breast cancer. Oncotarget 7:40189–40199. https://doi.org/10.18632/ONCOTARGET.9561
Khan S, Jutzy JMS, Valenzuela MMA et al (2012) Plasma-derived exosomal survivin, a plausible biomarker for early detection of prostate cancer. PLoS One 7. https://doi.org/10.1371/JOURNAL.PONE.0046737
Khan S, Bennit HF, Turay D et al (2014) Early diagnostic value of survivin and its alternative splice variants in breast cancer. BMC Cancer 14. https://doi.org/10.1186/1471-2407-14-176
Warmoes M, Lam SW, van der Groep P et al (2016) Secretome proteomics reveals candidate non-invasive biomarkers of BRCA1 deficiency in breast cancer. Oncotarget 7:63537–63548. https://doi.org/10.18632/ONCOTARGET.11535
Chanteloup G, Cordonnier M, Isambert N et al (2020) Monitoring HSP70 exosomes in cancer patients’ follow up: a clinical prospective pilot study. J Extracell Vesicles 9:1766192. https://doi.org/10.1080/20013078.2020.1766192
Min L, Zhu S, Chen L et al (2019) Evaluation of circulating small extracellular vesicles derived miRNAs as biomarkers of early colon cancer: a comparison with plasma total miRNAs. J Extracell Vesicles 8. https://doi.org/10.1080/20013078.2019.1643670/SUPPL_FILE/ZJEV_A_1643670_SM8506.ZIP
Ogata-Kawata H, Izumiya M, Kurioka D et al (2014) Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS One 9. https://doi.org/10.1371/JOURNAL.PONE.0092921
Bryant RJ, Pawlowski T, Catto JWF et al (2012) Changes in circulating microRNA levels associated with prostate cancer. Br J Cancer 106:768–774. https://doi.org/10.1038/BJC.2011.595
Matsuzaki K, Fujita K, **gushi K et al (2017) MiR-21-5p in urinary extracellular vesicles is a novel biomarker of urothelial carcinoma. Oncotarget 8:24668–24678. https://doi.org/10.18632/ONCOTARGET.14969
Armstrong DA, Green BB, Seigne JD et al (2015) MicroRNA molecular profiling from matched tumor and bio-fluids in bladder cancer. Mol Cancer 14. https://doi.org/10.1186/S12943-015-0466-2
Kim MW, Park S, Lee H et al (2021) Multi-miRNA panel of tumor-derived extracellular vesicles as promising diagnostic biomarkers of early-stage breast cancer. Cancer Sci. https://doi.org/10.1111/CAS.15155
Hannafon BN, Trigoso YD, Calloway CL et al (2016) Plasma exosome microRNAs are indicative of breast cancer. Breast Cancer Res 18. https://doi.org/10.1186/S13058-016-0753-X
Worst TS, Previti C, Nitschke K et al (2020) miR-10a-5p and miR-29b-3p as extracellular vesicle-associated prostate cancer detection markers. Cancers 12. https://doi.org/10.3390/CANCERS12010043
Asano N, Matsuzaki J, Ichikawa M et al (2019) A serum microRNA classifier for the diagnosis of sarcomas of various histological subtypes. Nat Commun 10:1–10. https://doi.org/10.1038/s41467-019-09143-8
Chevillet JR, Kang Q, Ruf IK et al (2014) Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc Natl Acad Sci U S A 111:14888–14893. https://doi.org/10.1073/pnas.1408301111
McKiernan J, Donovan MJ, O’Neill V et al (2016) A novel urine exosome gene expression assay to predict high-grade prostate cancer at initial biopsy. JAMA Oncol 2:882–889. https://doi.org/10.1001/JAMAONCOL.2016.0097
Peng Q, Chiu PKF, Wong CYP et al (2021) Identification of piRNA targets in urinary extracellular vesicles for the diagnosis of prostate cancer. Diagnostics (Basel) 11. https://doi.org/10.3390/DIAGNOSTICS11101828
Lu L, Fang S, Zhang Y et al (2021) Exosomes and exosomal circRNAs: the rising stars in the progression, diagnosis and prognosis of gastric cancer. Cancer Manag Res 13:8121–8129. https://doi.org/10.2147/CMAR.S331221
Zhao X, Guo X, Jiao D et al (2021) Analysis of the expression profile of serum exosomal lncRNA in breast cancer patients. Ann Transl Med 9:1382. https://doi.org/10.21037/ATM-21-3483
Thakur BK, Zhang H, Becker A et al (2014) Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res 24:766–769. https://doi.org/10.1038/cr.2014.44
Lee JS, Hur JY, Kim IA et al (2018) Liquid biopsy using the supernatant of a pleural effusion for EGFR genoty** in pulmonary adenocarcinoma patients: a comparison between cell-free DNA and extracellular vesicle-derived DNA. BMC Cancer 18:1–8. https://doi.org/10.1186/s12885-018-5138-3
Fernando MR, Jiang C, Krzyzanowski GD, Ryan WL (2017) New evidence that a large proportion of human blood plasma cell-free DNA is localized in exosomes. PLoS One 12:1–15. https://doi.org/10.1371/journal.pone.0183915
Wan Y, Liu B, Lei H et al (2018) Nanoscale extracellular vesicle-derived DNA is superior to circulating cell-free DNA for mutation detection in early-stage non-small-cell lung cancer. Ann Oncol 29:2379–2383. https://doi.org/10.1093/ANNONC/MDY458
Castellanos-Rizaldos E, Grimm DG, Tadigotla V et al (2018) Exosome-based detection of EGFR T790M in plasma from non-small cell lung cancer patients. Clin Cancer Res Off J Am Assoc Cancer Res 24:2944–2950. https://doi.org/10.1158/1078-0432.CCR-17-3369
Madhavan B, Yue S, Galli U et al (2015) Combined evaluation of a panel of protein and miRNA serum-exosome biomarkers for pancreatic cancer diagnosis increases sensitivity and specificity. Int J Cancer 136:2616–2627. https://doi.org/10.1002/IJC.29324
Brzozowski JS, Jankowski H, Bond DR et al (2018) Lipidomic profiling of extracellular vesicles derived from prostate and prostate cancer cell lines. Lipids Health Dis 17:1–12. https://doi.org/10.1186/S12944-018-0854-X/FIGURES/4
Zhang Y, Wu X, Tao WA (2018) Characterization and applications of extracellular vesicle proteome with post-translational modifications. Trends Anal Chem 107:21–30. https://doi.org/10.1016/J.TRAC.2018.07.014
Chen I-H, Xue L, Hsu C-C et al (2017) Phosphoproteins in extracellular vesicles as candidate markers for breast cancer. Proc Natl Acad Sci 114:3175–3180. https://doi.org/10.1073/pnas.1618088114
Luo P, Mao K, Xu J et al (2020) Metabolic characteristics of large and small extracellular vesicles from pleural effusion reveal biomarker candidates for the diagnosis of tuberculosis and malignancy. J Extracell Vesicles 9:1790158. https://doi.org/10.1080/20013078.2020.1790158
Gualerzi A, Picciolini S, Carlomagno C et al (2019) Raman profiling of circulating extracellular vesicles for the stratification of Parkinson’s patients. Nanomed Nanotechnol Biol Med 22. https://doi.org/10.1016/J.NANO.2019.102097
Enciso-Martinez A, Van Der Pol E, Hau CM et al (2020) Label-free identification and chemical characterisation of single extracellular vesicles and lipoproteins by synchronous Rayleigh and Raman scattering. J Extracell Vesicles 9. https://doi.org/10.1080/20013078.2020.1730134
Penders J, Nagelkerke A, Cunnane EM et al (2021) Single particle automated Raman trap** analysis of breast cancer cell-derived extracellular vesicles as cancer biomarkers. ACS Nano. https://doi.org/10.1021/acsnano.1c07075
LeClaire M, Gimzewski J, Sharma S (2021) A review of the biomechanical properties of single extracellular vesicles. Nano Select 2:1–15. https://doi.org/10.1002/nano.202000129
Barnes JM, Nauseef JT, Henry MD (2012) Resistance to fluid shear stress is a conserved biophysical property of malignant cells. PLoS One 7:e50973. https://doi.org/10.1371/journal.pone.0050973
Mitchell MJ, Denais C, Chan MF et al (2015) Lamin A/C deficiency reduces circulating tumor cell resistance to fluid shear stress. Am J Physiol Cell Physiol 309:C736–C746. https://doi.org/10.1152/ajpcell.00050.2015
Moose DL, Krog BL, Kim T-H et al (2020) Cancer cells resist mechanical destruction in circulation via RhoA/actomyosin-dependent mechano-adaptation. Cell Rep 30:3864–3874.e6. https://doi.org/10.1016/j.celrep.2020.02.080
Margolis E, Brown G, Partin A et al (2021) Predicting high-grade prostate cancer at initial biopsy: clinical performance of the ExoDx (EPI) prostate intelliscore test in three independent prospective studies. Prostate Cancer Prostatic Dis. https://doi.org/10.1038/S41391-021-00456-8
Ayers L, Pink R, Carter DRF, Nieuwland R (2019) Clinical requirements for extracellular vesicle assays. 8(1):1593755. https://doi.org/10.1080/20013078.2019.1593755
Coumans FAW, Brisson AR, Buzas EI et al (2017) Methodological guidelines to study extracellular vesicles. Circ Res 120:1632–1648. https://doi.org/10.1161/CIRCRESAHA.117.309417
Buscail E, Alix-Panabières C, Quincy P et al (2019) High clinical value of liquid biopsy to detect circulating tumor cells and tumor exosomes in pancreatic ductal adenocarcinoma patients eligible for up-front surgery. Cancers (Basel) 11:1656. https://doi.org/10.3390/CANCERS11111656
Suo Y, Gu Z, Wei X (2020) Advances of in vivo flow cytometry on cancer studies. Cytometry A 97:15–23. https://doi.org/10.1002/cyto.a.23851
Acknowledgments
We thank the Tumor Biomechanics lab (www.goetzlab.fr), for support and discussions. This work has been supported by Plan Cancer, INCa, Cancéropôle Grand-Est and La Ligue Contre le Cancer and by institutional funds from INSERM and the University of Strasbourg.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this chapter
Cite this chapter
Hyenne, V., Goetz, J.G., Osmani, N. (2022). Liquid Biopsies: Flowing Biomarkers. In: Caballero, D., Kundu, S.C., Reis, R.L. (eds) Microfluidics and Biosensors in Cancer Research. Advances in Experimental Medicine and Biology, vol 1379. Springer, Cham. https://doi.org/10.1007/978-3-031-04039-9_14
Download citation
DOI: https://doi.org/10.1007/978-3-031-04039-9_14
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-04038-2
Online ISBN: 978-3-031-04039-9
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)