Abstract
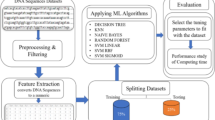
Information based on DNA-sequence is significant for scientists to examine the functionalities of genes. Recently, there is a tremendous increase in biological-data generation. Hence it is essential to match and classify the DNA sequences. This study aims to perform effective approaches for pre-processing and feature extraction of DNA data. Here the feature extraction is performed using the count vectorizer. Besides, it intended to classify the DNA sequences using machine learning (ML) algorithms using k-mer function. The matched sequences are retrieved effectively via the use of a pattern-matching algorithm. The results revealed that the SVM linear classifier gives good result.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Celli, F., Cumbo, F., Weitschek, E.: Classification of large DNA methylation datasets for identifying cancer drivers. Big Data Res. 13, 21–28 (2018)
Budach, S., Marsico, A.: Pysster: classification of biological sequences by learning sequence and structure motifs with convolutional neural networks. Bioinformatics 34, 3035–3037 (2018)
Phan, D., Ngoc, G.N., Lumbanraja, F.R., Faisal, M.R., Abipihi, B., Purnama, B., et al.: Combined use of k-mer numerical features and position-specific categorical features in fixed-length DNA sequence classification. J. Biomed. Sci. Eng. 10, 390–401 (2017)
Sahakyan, A.B., Chambers, V.S., Marsico, G., Santner, T., Di Antonio, M., Balasubramanian, S.: Machine learning model for sequence-driven DNA G-quadruplex formation. Sci. Rep. 7, 1–11 (2017)
Liu, B.: BioSeq-Analysis: a platform for DNA, RNA and protein sequence analysis based on machine learning approaches. Brief. Bioinform. 20, 1280–1294 (2019)
He, T., Jiao, L., Wiedenhoeft, A.C., Yin, Y.: Machine learning approaches outperform distance-and tree-based methods for DNA barcoding of Pterocarpus wood. Planta 249, 1617–1625 (2019)
Liu, B., Chen, S., Yan, K., Weng, F.: iRO-PsekGCC: identify DNA replication origins based on pseudo k-tuple GC composition. Front. Genet. 10, 842 (2019)
Aliferi, A., Ballard, D., Gallidabino, M.D., Thurtle, H., Barron, L., Court, D.S.: DNA methylation-based age prediction using massively parallel sequencing data and multiple machine learning models. Forensic Sci. Int. Genet. 37, 215–226 (2018)
Chowdhury, B., Garai, G.: A review on multiple sequence alignment from the perspective of genetic algorithm. Genomics 109, 419–431 (2017)
Tahir, M., Sardaraz, M., Ikram, A.A.: EPMA: efficient pattern matching algorithm for DNA sequences. Expert Syst. Appl. 80, 162–170 (2017)
Memeti, S., Pllana, S.: A machine learning approach for accelerating DNA sequence analysis. Int. J. High Perform. Comput. Appl. 32, 363–379 (2018)
Moghadam, B.T., Etemadikhah, M., Rajkowska, G., Stockmeier, C., Grabherr, M., Komorowski, J., et al.: Analyzing DNA methylation patterns in subjects diagnosed with schizophrenia using machine learning methods. J. Psychiatr. Res. 114, 41–47 (2019)
Touati, R., Messaoudi, I., Oueslati, A., Lachiri, Z., Kharrat, M.: New intraclass helitrons classification using DNA-image sequences and machine learning approaches. In: IRBM (2020)
Norlin, S.: DNA Sequence Classification Using Variable Length Markov Models (2020)
Ryu, C., Lecroq, T., Park, K.: Fast string matching for DNA sequences. Theoret. Comput. Sci. 812, 137–148 (2020)
Li, H., Xu, G., Tang, Q., Lin, X., Shen, X.S.: Enabling efficient and fine-grained DNA similarity search with access control over encrypted cloud data. In: International Conference on Wireless Algorithms, Systems, and Applications, pp. 236–248 (2018)
Anandakumar, M., Aiswarya, M.S., Bakyalakshmi, M.N., Brindha, M.S.: Pattern Similarity Search Using Expectation Maximization (Em) Algorithm
Xu, G., Li, H., Ren, H., Lin, X., Shen, X.S.: DNA similarity search with access control over encrypted cloud data. IEEE Trans. Cloud Comput. (2020)
Yin, B., Balvert, M., Zambrano, D., Schönhuth, A., Bohte, S.: An image representation based convolutional network for DNA classification, ar**v preprint ar**v:1806.04931 (2018)
Wilkinson, S.P., Davy, S.K., Bunce, M., Stat, M.: Taxonomic identification of environmental DNA with informatic sequence classification trees. PeerJ Preprints (2018)
Szalkaia, B., Grolmusza, V.: SECLAF: a webserver and deep neural network design tool for biological sequence classification, ar**v preprint ar**v:1708.04103 (2017)
Varsani, A., Krupovic, M.: Sequence-based taxonomic framework for the classification of uncultured single-stranded DNA viruses of the family Genomoviridae. Virus Evol. 3, vew037 (2017)
Touati, R., Oueslati, A.E., Messaoudi, I., Lachiri, Z.: The Helitron family classification using SVM based on Fourier transform features applied on an unbalanced dataset. Med. Biol. Eng. Compu. 57, 2289–2304 (2019)
Greenside, P., Shimko, T., Fordyce, P., Kundaje, A.: Discovering epistatic feature interactions from neural network models of regulatory DNA sequences. Bioinformatics 34, i629–i637 (2018)
Yang, A., Zhang, W., Wang, J., Yang, K., Han, Y., Zhang, L.: Review on the application of machine learning algorithms in the sequence data mining of DNA. Front. Bioeng. Biotechnol. 8, 1032 (2020)
Colbran, L.L., Chen, L., Capra, J.A.: Short DNA sequence patterns accurately identify broadly active human enhancers. BMC Genomics 18, 1–11 (2017)
Ravikumar, M., Prashanth, M.C.: Analysis of DNA sequence pattern matching: a brief survey. In: 2nd International Conference on Cybernetics, Cognition and Machine Learning Applications, (ICCCMLA 2020), Goa, pp. 221–229 (2020)
Ravikumar, M., Prashanth, M.C., Shivaprasad, B.J.: Searching pattern in DNA sequence using ECC-DiffieHellman exchange based hash function: an efficient approach. In: International Conference on Machine Learning and Big Data Analytics (ICMLBDA 2021), Patna (2021)
Neamatollahi, P., Hadi, M., Naghibzadeh, M.: Simple and efficient pattern matching algorithms for biological sequences. IEEE Access 8, 23838–23846 (2020)
Zhang, J., Bi, C., Wang, Y., Zeng, T., Liao, B., Chen, L.: Efficient mining closed k-mers from DNA and protein sequences. In: 2020 IEEE International Conference on Big Data and Smart Computing (BigComp), pp. 342–349 (2020)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this chapter
Cite this chapter
Ravikumar, M., Prashanth, M.C., Guru, D.S. (2022). Matching Pattern in DNA Sequences Using Machine Learning Approach Based on K-Mer Function. In: Gunjan, V.K., Zurada, J.M. (eds) Modern Approaches in Machine Learning & Cognitive Science: A Walkthrough. Studies in Computational Intelligence, vol 1027. Springer, Cham. https://doi.org/10.1007/978-3-030-96634-8_14
Download citation
DOI: https://doi.org/10.1007/978-3-030-96634-8_14
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-96633-1
Online ISBN: 978-3-030-96634-8
eBook Packages: Intelligent Technologies and RoboticsIntelligent Technologies and Robotics (R0)