Abstract
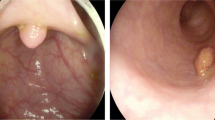
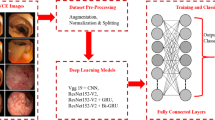
Polyp detection in wireless capsule endoscopy (WCE) is still an unsolved problem due to the large variation of polyps in terms of shape, color and size. There are two major problems hindering its improvement. First, traditional hand crafting approaches of WCE abnormalities’ detection has to be designed from scratch; it suffers from either a very time consuming process and/or a lack of exactitude. Second, WCE datasets acquisition still provides a challenge owing to the lack of large and publicly available annotated datasets. Recently, deep transfer learning has been widely used to transfer knowledge to medical images enabling the extraction of highly representative features. This paper investigates different architectures of pre-trained convolution neural networks (CNNs) from scratch (or network fine-tuning) for WCE polyp classification task. We compare the results with the state-of-art methods. The experiments consistently demonstrate that the use of a well-known DCNN architecture named Inception V3 with adequate fine-tuning outperform or, in the worst case, perform as a CNN trained from scratch. The last fully connected (fc) layer is connected to the support vector machine (SVM) classifier to obtain better accuracy. The methodology exceeds traditional handcrafting features extraction methods in terms of performance for WCE polyp abnormalities’ detection.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Souaidi, M., Charfi, S., Abdelouahad, A.A., El Ansari, M.: New features for wireless capsule endoscopy polyp detection. In: 2018 International Conference on Intelligent Systems and Computer Vision (ISCV), pp. 1–6. IEEE (2018)
Souaidi, M., Abdelouahad, A.A., El Ansari, M.: A fully automated ulcer detection system for wireless capsule endoscopy images. In: 2017 International Conference on Advanced Technologies for Signal and Image Processing (ATSIP), pp. 1–6. IEEE (2017)
Souaidi, M., Abdelouahed, A.A., El Ansari, M.: Multi-scale completed local binary patterns for ulcer detection in wireless capsule endoscopy images. Multimedia Tools Appl. 78(10), 13091–13108 (2019)
Rokkas, T., Papaxoinis, K., Triantafyllou, K., Ladas, S.D.: A meta-analysis evaluating the accuracy of colon capsule endoscopy in detecting colon polyps. Gastrointest. Endosc. 71(4), 792–798 (2010)
Li, B., Meng, M.Q.-H.: Automatic polyp detection for wireless capsule endoscopy images. Exp. Syst. Appl. 39(12), 10952–10958 (2012)
Yuan, Y., Li, B., Meng, M.Q.-H.: Improved bag of feature for automatic polyp detection in wireless capsule endoscopy images. IEEE Trans. Autom. Sci. Eng. 13(2), 529–535 (2016)
Charfi, S., El Ansari, M.: Computer-aided diagnosis system for colon abnormalities detection in wireless capsule endoscopy images. Multimedia Tools Appl. 77(3), 4047–4064 (2018)
Shin, H.-C., Lu, L., Kim, L., Seff, A., Yao, J., Summers, R.M.: Interleaved text/image deep mining on a very large-scale radiology database. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 1090–1099 (2015)
Bernal, J., et al.: Comparative validation of polyp detection methods in video colonoscopy: results from the MICCAI 2015 endoscopic vision challenge. IEEE Trans. Med. Imaging 36(6), 1231–1249 (2017)
Tajbakhsh, N., et al.: Convolutional neural networks for medical image analysis: full training or fine tuning? IEEE Trans. Med. Imaging 35(5), 1299–1312 (2016)
Bartler, A., Mauch, L., Yang, B., Reuter, M., Stoicescu, L.: Automated detection of solar cell defects with deep learning. In: 26th European Signal Processing Conference (EUSIPCO), vol. 2018, pp. 2035–2039. IEEE (2018)
Szegedy, C., et al.: Going deeper with convolutions. In: 2015 Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 1–9 (2015)
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: 2016 Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 770–778 (2016)
Aoki, T., et al.: Automatic detection of erosions and ulcerations in wireless capsule endoscopy images based on a deep convolutional neural network. Gastrointest. Endosc. 89(2), 357–363 (2019)
Seguí, S., et al.: Deep learning features for wireless capsule endoscopy analysis. In: Beltrán-Castañón, C., Nyström, I., Famili, F. (eds.) CIARP 2016. LNCS, vol. 10125, pp. 326–333. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-52277-7_40
Jia, X., Meng, M.Q.-H.: Gastrointestinal bleeding detection in wireless capsule endoscopy images using handcrafted and CNN features. In: 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), vol. 2017, pp. 3154–3157. IEEE (2017)
Chatfield, K., Simonyan, K., Vedaldi, A., Zisserman, A.: Return of the devil in the details: delving deep into convolutional nets. ar**v preprint ar**v:1405.3531 (2014)
Mash, R., Borghetti, B., Pecarina, J.: Improved aircraft recognition for aerial refueling through data augmentation in convolutional neural networks. In: Bebis, G., et al. (eds.) ISVC 2016. LNCS, vol. 10072, pp. 113–122. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-50835-1_11
O’Shea, K., Nash, R.: An introduction to convolutional neural networks. ar**v preprint ar**v:1511.08458 (2015)
Kavukcuoglu, K., Sermanet, P., Boureau, Y.-L., Gregor, K., Mathieu, M., Cun, Y.L.: Learning convolutional feature hierarchies for visual recognition. In: 2010 Advances in Neural Information Processing Systems, pp. 1090–1098 (2010)
Hinton, G.E., Osindero, S., Teh, Y.-W.: A fast learning algorithm for deep belief nets. Neural Comput. 18(7), 1527–1554 (2006)
Wu, H., Gu, X.: Max-pooling dropout for regularization of convolutional neural networks. In: Arik, S., Huang, T., Lai, W.K., Liu, Q. (eds.) ICONIP 2015. LNCS, vol. 9489, pp. 46–54. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-26532-2_6
Deng, J., Dong, W., Socher, R., Li, L.-J., Li, K., Fei-Fei, L.: ImageNet: a large-scale hierarchical image database. In: IEEE Conference on Computer Vision and Pattern Recognition, vol. 2009, pp. 248–255. IEEE (2009)
Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J., Wojna, Z.: Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, vol. 2016, pp. 2818–2826 (2016)
Esteva, A., et al.: Dermatologist-level classification of skin cancer with deep neural networks. Nature 542(7639), 115 (2017)
Wang, L.: Support Vector Machines: Theory and Applications, vol. 177. Springer, Heidelberg (2005). https://doi.org/10.1007/b95439
WEO: WEO clinical endoscopy Atlas (1962). http://www.endoatlas.org/
Picard, R.R., Cook, R.D.: Cross-validation of regression models. J. Am. Stat. Assoc. 79(387), 575–583 (1984)
Peng, C.-Y.J., Lee, K.L., Ingersoll, G.M.: An introduction to logistic regression analysis and reporting. J. Educ. Res. 96(1), 3–14 (2002)
Sainath, T.N., Vinyals, O., Senior, A., Sak, H.: Convolutional, long short-term memory, fully connected deep neural networks. In: 2015 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), vol. 2015, pp. 4580–4584. IEEE (2015)
Pedregosa, F., et al.: Scikit-learn: machine learning in Python, pp. 2825–2830 (2011). https://scikit-learn.org/stable/modules/generated/ sklearn.model_selection.GridSearchCV.html/
Pogorelov, K., et al.: KVASIR: a multi-class image dataset for computer aided gastrointestinal disease detection. In: Proceedings of the 8th ACM on Multimedia Systems Conference, vol. 2017, pp. 164–169. ACM (2017)
Tindall, L., Luong, C., Saad, A.: Plankton classification using VGG16 network (2015)
Guo, Y., Liu, Y., Oerlemans, A., Lao, S., Wu, S., Lew, M.S.: Deep learning for visual understanding: a review. Neurocomputing 187, 27–48 (2016)
Goodfellow, I., et al.: Generative adversarial nets. In: Advances in Neural Information Processing Systems, vol. 2014, pp. 2672–2680 (2014)
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Souaidi, M., El Ansari, M. (2022). Automated Detection of Wireless Capsule Endoscopy Polyp Abnormalities with Deep Transfer Learning and Support Vector Machines. In: Kacprzyk, J., Balas, V.E., Ezziyyani, M. (eds) Advanced Intelligent Systems for Sustainable Development (AI2SD’2020). AI2SD 2020. Advances in Intelligent Systems and Computing, vol 1417. Springer, Cham. https://doi.org/10.1007/978-3-030-90633-7_74
Download citation
DOI: https://doi.org/10.1007/978-3-030-90633-7_74
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-90632-0
Online ISBN: 978-3-030-90633-7
eBook Packages: Intelligent Technologies and RoboticsIntelligent Technologies and Robotics (R0)