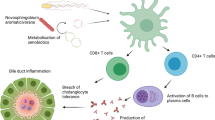
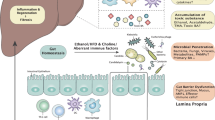
Abstract
Interactions between the gut and the liver in relation to immunological responses and metabolic functions have been recognized as central to homeostasis for decades. The roles of gut bacteria in the pathogenesis of certain complications of cirrhosis such as hepatic encephalopathy, spontaneous bacterial peritonitis, and systemic sepsis were also delineated decades ago – what is new is the idea that interactions between luminal bacteria in the gut (microbiota), the gut itself, and the liver may play a more fundamental role in the pathogenesis of a number of metabolic and immunological diseases. From basic animal research, as well as observations in human disease, a general hypothesis has emerged to explain how interplay between these factors might initiate or perpetuate alcoholic liver disease, nonalcoholic fatty liver disease, and even immunologically mediated liver diseases. In this framework, disrupted microbiota and their products gain access to the gut-associated immune system via a permeable gut barrier and generate inflammatory responses. Disruption of the gut vascular barrier then permits access for bacterial components as well as pro-inflammatory cytokines to the portal circulation and onto the liver where inflammatory and metabolic responses drive the genesis of various liver diseases. Cross-reactivity between bacterial antigens and biliary epitopes has also been invoked in relation to primary sclerosing cholangitis and primary biliary cholangitis. As we learn more of the details of these interactions and of the primacy, or otherwise, of the microbiota in this microbiota-gut-liver axis, new preventive strategies and therapeutic avenues may open up.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Simbrunner B, Mandorfer M, Trauner M, Reiberger T. Gut-liver axis signaling in portal hypertension. World J Gastroenterol. 2019;25:5897–917.
Broun GO, McMaster PD, Rous P. Studies on the total bile: IV. The enterohepatic circulation of bile pigment. J Exp Med. 1923;37:699–710.
Dowling RH, Mack E, Small DM. Effects of controlled interruption of the enterohepatic circulation of bile salts by biliary diversion and by ileal resection on bile salt secretion, synthesis, and pool size in the rhesus monkey. J Clin Invest. 1970;49:232–42.
Dowling RH. The enterohepatic circulation. Gastroenterology. 1972;62:122–40.
Emery FE, Joyce HE. Enterohepatic circulation of oestrogens. J Endocrinol. 1946;4:371–4.
Briggs FN, Taurog A, Chaikoff IL. The enterohepatic circulation of thyroxine in the rat. Endocrinology. 1953;52:559–67.
Lester R, Ostrow JD, Schmid R. Enterohepatic circulation of bilirubin. Nature. 1961;192:372.
Di Ciaula A, Garruti G, Lunardi Baccetto R, Molina-Molina E, Bonfrate L, Wang DQ, Portincasa P. Bile acid physiology. Ann Hepatol. 2017;16(Suppl. 1: s3–105):s4–s14.
Davis BC, Bajaj JS. The human gut microbiome in liver diseases. Semin Liver Dis. 2017;37:128–40.
Phillips GB, Schwartz R, Gabuzda GJ Jr, Davidson CS. The syndrome of impending hepatic coma in patients with cirrhosis of the liver given certain nitrogenous substances. N Engl J Med. 1952;247:239–46.
Martini GA, Phear EA, Ruebner B, Sherlock S. The bacterial content of the small intestine in normal and cirrhotic subjects: relation to methionine toxicity. Clin Sci. 1957;16:35–51.
Phear EA, Ruebner B, Sherlock S, Summerskill WH. Methionine toxicity in liver disease and its prevention by chlortetracycline. Clin Sci. 1956;15:93–117.
Quigley EMM. Gastrointestinal dysfunction in liver disease – gut-liver interactions revisited. Dig Dis Sci. 1996;41:557–61.
Thalheimer U, Triantos CK, Samonakis DN, Patch D, Burroughs AK. Infection, coagulation and variceal bleeding in cirrhosis. Gut. 2005;54:556–63.
Quigley EM, Abu-Shanab A, Murphy EF, Stanton C, Monsour HP Jr. The metabolic role of the microbiome: implications for NAFLD and the metabolic syndrome. Semin Liver Dis. 2016;36:312–6.
Brandl K, Kumar V, Eckmann L. Gut-liver axis at the frontier of host-microbial interactions. Am J Physiol Gastrointest Liver Physiol. 2017;312:G413–9.
Arab JP, Martin-Mateos RM, Shah VH. Gut-liver axis, cirrhosis and portal hypertension: the chicken and the egg. Hepatol Int. 2018;12(Suppl 1):24–33.
Tripathi A, Debelius J, Brenner DA, Karin M, Loomba R, Schnabl B, Knight R. The gut-liver axis and the intersection with the microbiome. Nat Rev Gastroenterol Hepatol. 2018;15:397–411.
Wiest R, Albillos A, Trauner M, Bajaj JS, Jalan R. Targeting the gut-liver axis in liver disease. J Hepatol. 2017;67:1084–103.
Safari Z, Gérard P. The links between the gut microbiome and non-alcoholic fatty liver disease (NAFLD). Cell Mol Life Sci. 2019;76:1541–58.
Quigley EM. Primary biliary cirrhosis and the microbiome. Semin Liver Dis. 2016;36:349–53.
O’Hara SP, LaRusso NF. The gut-liver axis in primary sclerosing cholangitis: are pathobionts the missing link? Hepatology. 2019;70:1058–60.
About the Human Microbiome. NIH Human Microbiome Project – About the Human Microbiome. https://hmpdacc.org/hmp/overview/. Accessed 2 Feb 2020.
Eloe-Fadrosh EA, Rasko DA. The human microbiome: from symbiosis to pathogenesis. Annu Rev Med. 2013;64:145–63.
Huang YJ, Boushey HA. The microbiome in asthma. J Allergy Clin Immunol. 2015;135:25–30.
Costello ME, Robinson PC, Benham H, Brown MA. The intestinal microbiome in human disease and how it relates to arthritis and spondyloarthritis. Best Pract Res Clin Rheumatol. 2015;29:202–12.
Mathur R, Barlow GM. Obesity and the microbiome. Expert Rev Gastroenterol Hepatol. 2015;9:1087–99.
Tang WH, Kitai T, Hazen SL. Gut microbiota in cardiovascular health and disease. Circ Res. 2017;120:1183–96.
Fung TC, Olson CA, Hsiao EY. Interactions between the microbiota, immune and nervous systems in health and disease. Nat Neurosci. 2017;20:145–55.
Lynch SV, Pedersen O. The human intestinal microbiome in health and disease. N Engl J Med. 2016;375:2369–79.
Claesson MJ, Clooney AG, O’Toole PW. A clinician’s guide to microbiome analysis. Nat Rev Gastroenterol Hepatol. 2017;14:585–95.
Collado MC, Rautava S, Aakko J, Isolauri E, Salminen S. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci Rep. 2016;6:23129.
Leiby JS, McCormick K, Sherrill-Mix S, et al. Lack of detection of a human placenta microbiome in samples from preterm and term deliveries. Microbiome. 2018;6(1):196.
Neu J. The microbiome during pregnancy and early postnatal life. Semin Fetal Neonatal Med. 2016;21:373–9.
Jakobsson HE, Abrahamsson TR, Jenmalm MC, Harris K, Quince C, Jernberg C, et al. Decreased gut microbiota diversity, delayed Bacteroidetes colonisation and reduced Th1 responses in infants delivered by caesarean section. Gut. 2014;63:559–66.
Dogra S, Sakwinska O, Soh SE, Ngom-Bru C, Brück WM, Berger B, Brüssow H, Lee YS, Yap F, Chong YS, Godfrey KM, Holbrook JD, GUSTO Study Group. Dynamics of infant gut microbiota are influenced by delivery mode and gestational duration and are associated with subsequent adiposity. mBio. 2015;6:e02419–4.
Cong X, Xu W, Janton S, Henderson WA, Matson A, McGrath JM, et al. Gut microbiome developmental patterns in early life of preterm infants: impacts of feeding and gender. PLoS One. 2016;11:e0152751.
Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. 2015;17:690–703.
Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–7.
Pannaraj PS, Li F, Cerini C, et al. Association between breast milk bacterial communities and establishment and development of the infant gut microbiome. JAMA Pediatr. 2017;171:647–54.
Tun HM, Bridgman SL, Chari R, et al. Roles of birth mode and infant gut microbiota in intergenerational transmission of overweight and obesity from mother to offspring. JAMA Pediatr. 2018;172:368–77.
Lundgren SN, Madan JC, Emond JA, Morrison HG, Christensen BC, Karagas MR, et al. Maternal diet during pregnancy is related with the infant stool microbiome in a delivery mode-dependent manner. Microbiome. 2018;6:109.
Cho I, Yamanishi S, Cox L, Methé BA, Zavadil J, Li K, et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature. 2012;488:621–6.
Cox LM, Yamanishi S, Sohn J, Alekseyenko AV, Leung JM, Cho I, et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell. 2014;158:705–21.
Dawson-Hahn EE, Rhee KE. The association between antibiotics in the first year of life and child growth trajectory. BMC Pediatr. 2019;19:23.
Saari A, Virta LJ, Sankilampi U, Dunkel L, Saxen H. Antibiotic exposure in infancy and risk of being overweight in the first 24 months of life. Pediatrics. 2015;135(4):617–26.
Poulsen MN, Pollak J, Bailey-Davis L, Hirsch AG, Glass TA, Schwartz BS. Associations of prenatal and childhood antibiotic use with child body mass index at age 3 years. Obesity (Silver Spring). 2017;25:438–44.
Block JP, Bailey LC, Gillman MW, Lunsford D, Daley MF, Eneli I, et al. Early antibiotic exposure and weight outcomes in young children. Pediatrics. 2018;142:e20180290.
Diaz Heijtz R. Fetal, neonatal, and infant microbiome: perturbations and subsequent effects on brain development and behavior. Semin Fetal Neonatal Med. 2016;21:410–7.
Forbes JD, Azad MB, Vehling L, Tun HM, Konya TB, Guttman DS, Canadian Healthy Infant Longitudinal Development (CHILD) Study Investigators, et al. Association of exposure to formula in the hospital and subsequent infant feeding practices with gut microbiota and risk of overweight in the first year of life. JAMA Pediatr. 2018;172:e181161.
Tun HM, Bridgman SL, Chari R, Field CJ, Guttman DS, Becker AB, Canadian Healthy Infant Longitudinal Development (CHILD) Study Investigators, et al. Roles of birth mode and infant gut microbiota in intergenerational transmission of overweight and obesity from mother to offspring. JAMA Pediatr. 2018;172:368–77.
Chelimo C, Camargo CA Jr, Morton SMB, Grant CC. Association of repeated antibiotic exposure up to age 4 years with body mass at age 4.5 years. JAMA Netw Open. 2020;3:e1917577.
Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–80.
Santoro A, Ostan R, Candela M, Biagi E, Brigidi P, Capri M, Franceschi C. Gut microbiota changes in the extreme decades of human life: a focus on centenarians. Cell Mol Life Sci. 2018;75:129–48.
Kumar M, Babaei P, Ji B, Nielsen J. Human gut microbiota and healthy aging: recent developments and future prospective. Nutr Healthy Aging. 2016;4:3–16.
Claesson MJ, Jeffery IB, Conde S, Power SE, O’Connor EM, Cusack S, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488:178–84.
Odamaki T, Kato K, Sugahara H, Hashikura N, Takahashi S, **ao JZ, et al. Age-related changes in gut microbiota composition from newborn to centenarian: a cross-sectional study. BMC Microbiol. 2016;16:90.
Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474:327–36.
Surana NK, Kasper DL. Deciphering the tête-à-tête between the microbiota and the immune system. J Clin Invest. 2014;124:4197–203.
Wells JM, Brummer RJ, Derrien M, MacDonald TT, Troost F, Cani PD, et al. Homeostasis of the gut barrier and potential biomarkers. Am J Physiol Gastrointest Liver Physiol. 2017;312:G171–93.
Dey N, Wagner VE, Blanton LV, Cheng J, Fontana L, Haque R, et al. Regulators of gut motility revealed by a gnotobiotic model of diet-microbiome interactions related to travel. Cell. 2015;163:95–107.
Kabouridis PS, Lasrado R, McCallum S, Chng SH, Snippert HJ, Clevers H, et al. The gut microbiota keeps enteric glial cells on the move; prospective roles of the gut epithelium and immune system. Gut Microbes. 2015;6:398–403.
Savidge TC. Epigenetic regulation of enteric neurotransmission by gut bacteria. Front Cell Neurosci. 2016;9:503.
Turnbaugh PJ, Gordon JI. The core gut microbiome, energy balance and obesity. J Physiol. 2009;587:4153–8.
Carmody RN, Turnbaugh PJ. Host-microbial interactions in the metabolism of therapeutic and diet-derived xenobiotics. J Clin Invest. 2014;124:4173–81.
Vonk RJ, Reckman G. Progress in the biology and analysis of short chain fatty acids. J Physiol. 2017;595:419–20.
Yano JM, Yu K, Donaldson GP, Shastri GG, Ann P, Ma L, et al. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell. 2015;161:264–76.
Wilson ID, Nicholson JK. Gut microbiome interactions with drug metabolism, efficacy and toxicity. Transl Res. 2017;179:204–22.
Shanahan F, van Sinderen D, O’Toole PW, Stanton C. Feeding the microbiota: transducer of nutrient signals for the host. Gut. 2017;66:1709–17.
Smith MI, Yatsunenko T, Manary MJ, Trehan I, Mkakosya R, Cheng J, et al. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science. 2013;339:548–54.
Subramanian S, Huq S, Yatsunenko T, Haque R, Mahfuz M, Alam MA, et al. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature. 2014;510:417–21.
Sonnenburg ED, Sonnenburg JL. Starving our microbial self: the deleterious consequences of a diet deficient in microbiota-accessible carbohydrates. Cell Metab. 2014;20:779–86.
McIntosh K, Reed DE, Schneider T, Dang F, Keshteli AH, De Palma G, et al. FODMAPs alter symptoms and the metabolome of patients with IBS: a randomised controlled trial. Gut. 2017;66(7):1241–51.
Clarke SF, Murphy EF, O’Sullivan O, Lucey AJ, Humphreys M, Hogan A, et al. Exercise and associated dietary extremes impact on gut microbial diversity. Gut. 2014;63:1913–20.
Hildebrandt MA, Hoffmann C, Sherrill-Mix SA, Keilbaugh SA, Hamady M, Chen YY, et al. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology. 2009;137:1716–24.
Heinritz SN, Weiss E, Eklund M, Aumiller T, Louis S, Rings A, et al. Intestinal microbiota and microbial metabolites are changed in a pig model fed a high-fat/low-fiber or a low-fat/high-fiber diet. PLoS One. 2016;11:e0154329.
Kovatcheva-Datchary P, Nilsson A, Akrami R, Lee YS, De Vadder F, Arora T, et al. Dietary fiber-induced improvement in glucose metabolism is associated with increased abundance of Prevotella. Cell Metab. 2015;22:971–82.
Sonnenburg ED, Smits SA, Tikhonov M, Higginbottom SK, Wingreen NS, Sonnenburg JL. Diet-induced extinctions in the gut microbiota compound over generations. Nature. 2016;529:212–5.
Degnan PH, Taga ME, Goodman AL. Vitamin B12 as a modulator of gut microbial ecology. Cell Metab. 2014;20:769–78.
Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–8.
Halmos EP, Christophersen CT, Bird AR, Shepherd SJ, Gibson PR, Muir JG. Diets that differ in their FODMAP content alter the colonic luminal microenvironment. Gut. 2015;64:93–100.
Bonder MJ, Tigchelaar EF, Cai X, Trynka G, Cenit MC, Hrdlickova B, et al. The influence of a short-term gluten-free diet on the human gut microbiome. Genome Med. 2016;8:45.
David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–63.
Luca F, Kupfer SS, Knights D, Khoruts A, Blekhman R. Functional genomics of host-microbiome interactions in humans. Trends Genet. 2017;34:30–40.
Dong TS, Gupta A. Influence of early life, diet, and the environment on the microbiome. Clin Gastroenterol Hepatol. 2019;17(2):231–42.
Mayer EA. Gut feelings: the emerging biology of gut-brain communication. Nat Rev Neurosci. 2011;12:453–66.
Modi SR, Collins JJ, Relman DA. Antibiotics and the gut microbiota. J Clin Invest. 2014;124:4212–8.
Blaser MJ. Antibiotic use and its consequences for the normal microbiome. Science. 2016;352:544–5.
Freedberg DE, Toussaint NC, Chen SP, Ratner AJ, Whittier S, Wang TC, Wang HH, Abrams JA. Proton pump inhibitors alter specific taxa in the human gastrointestinal microbiome: a crossover trial. Gastroenterology. 2015;149:883–5.
Jackson MA, Goodrich JK, Maxan ME, Freedberg DE, Abrams JA, Poole AC, et al. Proton pump inhibitors alter the composition of the gut microbiota. Gut. 2016;65:749–56.
Forslund K, Hildebrand F, Nielsen T, Falony G, Le Chatelier E, Sunagawa S, et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature. 2015;528:262–6.
Quigley EMM. Gut microbiome as a clinical tool in gastrointestinal disease management: are we there yet? Nat Rev Gastroenterol Hepatol. 2017;14:315–20.
Hoefert B. Über die Bakterienbefunde im Duodenalsaft von Gesunden und Kranken. Zschr Klin Med. 1921;92:221–35.
Shah A, Shanahan E, Macdonald GA, Fletcher L, Ghasemi P, Morrison M, et al. Systematic review and meta-analysis: prevalence of small intestinal bacterial overgrowth in chronic liver disease. Semin Liver Dis. 2017;37:388–400.
Maslennikov R, Pavlov C, Ivashkin V. Small intestinal bacterial overgrowth in cirrhosis: systematic review and meta-analysis. Hepatol Int. 2018;12:567–76.
Augustyn M, Grys I, Kukla M. Small intestinal bacterial overgrowth and nonalcoholic fatty liver disease. Clin Exp Hepatol. 2019;5:1–10.
Ghosh G, Jesudian AB. Small intestinal bacterial overgrowth in patients with cirrhosis. J Clin Exp Hepatol. 2019;9:257–67.
Liu Chen Kiow J, Vincent C, Sidani S, Bouin M. High occurrence of small intestinal bacterial overgrowth in primary biliary cholangitis. Neurogastroenterol Motil. 2019;31:e13691.
Wijarnpreecha K, Lou S, Watthanasuntorn K, Kroner PT, Cheungpasitporn W, Lukens FJ, et al. Small intestinal bacterial overgrowth and nonalcoholic fatty liver disease: a systematic review and meta-analysis. Eur J Gastroenterol Hepatol. 2020;32(5):601–8.
Bode C, Kolepke R, Schafer K, Bode JC. Breath hydrogen excretion in patients with alcoholic liver disease–evidence of small intestinal bacterial overgrowth. Z Gastroenterol. 1993;31:3–7.
Teltschik Z, Wiest R, Beisner J, Nuding S, Hofmann C, Schoelmerich J, et al. Intestinal bacterial translocation in rats with cirrhosis is related to compromised Paneth cell antimicrobial host defense. Hepatology. 2012;55:1154–63.
Purohit V, Bode JC, Bode C, Brenner DA, Choudhry MA, Hamilton F, et al. Alcohol, intestinal bacterial growth, intestinal permeability to endotoxin, and medical consequences: summary of a symposium. Alcohol. 2008;42:349–61.
Bajaj JS. Alcohol, liver disease and the gut microbiota. Nat Rev Gastroenterol Hepatol. 2019;16:235–46.
Wigg AJ, Roberts-Thomson IC, Dymock RB, McCarthy PJ, Grose RH, Cumming AG. The role of small intestinal bacterial overgrowth, intestinal permeability, endotoxemia, and tumor necrosis factor alpha in the pathogenesis of nonalcoholic steatohepatitis. Gut. 2001;48:206–11.
Abu Shanab A, Scully P, Crosbie O, Buckley M, O’Mahony L, Shanahan F, et al. Small intestinal bacterial overgrowth in non-alcoholic steato-hepatitis; association with toll-like receptor 4 expression and plasma levels of interleukin 8. Dig Dis Sci. 2011;56:1524–34.
Kolodziejczyk AA, Zheng D, Shibolet O, Elinav E. The role of the microbiome in NAFLD and NASH. EMBO Mol Med. 2019;11:pii: e9302.
Vanderhoof JA, Tuma DJ, Antonson DL, Sorrell MF. Effect of antibiotics in the prevention of jejunoileal bypass-induced liver dysfunction. Digestion. 1982;23:9–15.
Quigley EMM. The spectrum of small intestinal bacterial overgrowth (SIBO). Curr Gastroenterol Rep. 2019;21:3.
Kastl AJ Jr, Terry NA, Albenberg LG, Wu GD. The structure and function of the human small intestinal microbiota: current understanding and future directions. Cell Mol Gastroenterol Hepatol. 2020;9:33–45.
Quigley EMM. Symptoms and the small intestinal microbiome – the unknown explored. Nat Rev Gastroenterol Hepatol. 2019;16:457–8.
Bajaj JS, Hylemon PB, Ridlon JM, Heuman DM, Daita K, White MB, et al. Colonic mucosal microbiome differs from stool microbiome in cirrhosis and hepatic encephalopathy and is linked to cognition and inflammation. Am J Physiol Gastrointest Liver Physiol. 2012;303:G675–85.
Chen Y, Yang F, Lu H, Wang B, Chen Y, Lei D, Wang Y, Zhu B, Li L. Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology. 2011;54:562–72.
Bajaj JS, Ridlon JM, Hylemon PB, Thacker LR, Heuman DM, Smith S, et al. Linkage of gut microbiome with cognition in hepatic encephalopathy. Am J Physiol Gastrointest Liver Physiol. 2012;302:G168–75.
Lu H, Wu Z, Xu W, Yang J, Chen Y, Li L. Intestinal microbiota was assessed in cirrhotic patients with hepatitis B virus infection. Intestinal microbiota of HBV cirrhotic patients. Microb Ecol. 2011;61:693–703.
Liu J, Wu D, Ahmed A, Li X, Ma Y, Tang L, Mo D, Ma Y, **n Y. Comparison of the gut microbe profiles and numbers between patients with liver cirrhosis and healthy individuals. Curr Microbiol. 2012;65:7–13.
Machado MV, Cortez-Pinto H. Gut microbiota and nonalcoholic fatty liver disease. Ann Hepatol. 2012;11:440–9.
Chen X, Devaraj S. Gut microbiome in obesity, metabolic syndrome, and diabetes. Curr Diab Rep. 2018;18:129.
Cani PD. Microbiota and metabolites in metabolic diseases. Nat Rev Endocrinol. 2019;15:69–70.
Canfora EE, Meex RCR, Venema K, Blaak EE. Gut microbial metabolites in obesity, NAFLD and T2DM. Nat Rev Endocrinol. 2019;15:261–73.
Greenblum S, Turnbaugh PJ, Borenstein E. Metagenomic systems biology of the human gut microbiome reveals topological shifts associated with obesity and inflammatory bowel disease. PNAS. 2012;109:594–9.
Karlsson FH, Tremaroli V, Nookaew I, Bergström G, Behre CJ, Fagerberg B, et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498:99–103.
Zupancic ML, Cantarel BL, Liu Z, Drabek EF, Ryan KA, Cirimotich S, et al. Analysis of the gut microbiota in the old order Amish and its relation to the metabolic syndrome. PLoS One. 2012;7:e43052.
Penas-Steinhardt A, Barcos LS, Belforte FS, de Sereday M, Vilarino J, Gonzalez CD, et al. Functional characterization of TLR4 +3725G/C polymorphism and association with protection against overweight. PLoS One. 2012;7:e50992.
Kim K-A, Gu W, Lee I-A, Joh E-H, Kim D-H. High fat diet-induced gut microbiota exacerbates inflammation and obesity in mice via the TLR4 signaling pathway. PLoS One. 2012;7:347713.
Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, et al. Metabolic syndrome and altered gut microbiota in mice lacking toll-like receptor 5. Science. 2010;328:228–31.
Fei N, Bruneau A, Zhang X, Wang R, Wang J, Rabot S, et al. Endotoxin producers overgrowing in human gut microbiota as the causative agents for nonalcoholic fatty liver disease. mBio. 2020;11:pii: e03263-19.
Schwimmer JB, Johnson JS, Angeles JE, Behling C, Belt PH, Borecki I, et al. Microbiome signatures associated with steatohepatitis and moderate to severe fibrosis in children with nonalcoholic fatty liver disease. Gastroenterology. 2019;157:1109–22.
Henao-Mejia J, Elinav E, ** C, Hao L, Mehal WZ, Strowig T, et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2012;482:179–85.
Tilg H, Moschen AR, Szabo G. Interleukin-1 and inflammasomes in alcoholic liver disease/acute alcoholic hepatitis and nonalcoholic fatty liver disease/nonalcoholic steatohepatitis. Hepatology. 2016;64:955–65.
Meroni M, Longo M, Dongiovanni P. Alcohol or gut microbiota: who is the guilty? Int J Mol Sci. 2019;20:pii: E4568.
Yuan J, Chen C, Cui J, Lu J, Yan C, Wei X, et al. Fatty liver disease caused by high-alcohol-producing Klebsiella pneumoniae. Cell Metab. 2019;30:675–88.
Zhu Y, Li F, Guo GL. Tissue-specific function of farnesoid X receptor in liver and intestine. Pharmacol Res. 2011;63:259–65.
Porez G, Prawitt J, Gross B, et al. Bile acid receptors as targets for the treatment of dyslipidemia and cardiovascular disease. J Lipid Res. 2012;53:1723–37.
Auwerx J, Messaddeq N, Sato H, Kodama T, Watanabe M, Ezaki O, et al. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature. 2006;439(7075):484–9.
Zhang Y, Lee FY, Lee H, Barrera G, Vales C, Gonzalez FJ, et al. Activation of the nuclear receptor FXR improves hyperglycemia and hyperlipidemia in diabetic mice. Proc Natl Acad Sci. 2006;103(4):1006–11.
Sorribas M, Jakob MO, Yilmaz B, Li H, Stutz D, Noser Y, et al. FXR modulates the gut-vascular barrier by regulating the entry sites for bacterial translocation in experimental cirrhosis. J Hepatol. 2019;71:1126–40.
Long SL, Gahan CGM, Joyce SA. Interactions between gut bacteria and bile in health and disease. Mol Asp Med. 2017;56:54–65.
Jia W, **e G, Jia W. Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat Rev Gastroenterol Hepatol. 2018;15:111–28.
Chen ML, Takeda K, Sundrud MS. Emerging roles of bile acids in mucosal immunity and inflammation. Mucosal Immunol. 2019;12:851–61.
Thanissery R, Winston JA, Theriot CM. Inhibition of spore germination, growth, and toxin activity of clinically relevant C. difficile strains by gut microbiota derived secondary bile acids. Anaerobe. 2017;45:86–100.
Jones BV, Begley M, Hill C, Gahan CG, Marchesi JR. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc Natl Acad Sci U S A. 2008;105:13580–5.
Vetrano S, Rescigno M, Cera MR, et al. Unique role of junctional adhesion molecule-a in maintaining mucosal homeostasis in inflammatory bowel disease. Gastroenterology. 2008;135:173–84.
Gunzel D, Yu AS. Claudins and the modulation of tight junction permeability. Physiol Rev. 2013;93:525–69.
Takiishi T, Fenero CIM, Câmara NOS. Intestinal barrier and gut microbiota: sha** our immune responses throughout life. Tissue Barriers. 2017;5:e1373208.
Camilleri M. Leaky gut: mechanisms, measurement and clinical implications in humans. Gut. 2019;68:1516–26.
Blaschitz C, Raffatellu M. Th17 cytokines and the gut mucosal barrier. J Clin Immunol. 2010;30:196–203.
Pontarollo G, Mann A, Brandão I, Malinarich F, Schöpf M, Reinhardt C. Protease-activated receptor signaling in intestinal permeability regulation. FEBS J. 2020;287(4):645–58.
Nicoletti A, Ponziani FR, Biolato M, Valenza V, Marrone G, Sgagna G, et al. Intestinal permeability in the pathogenesis of liver damage: from non-alcoholic fatty liver disease to liver transplantation. World J Gastroenterol. 2019;25:4814–34.
Munford RS. Endotoxemia-menace, marker, or mistake? J Leukoc Biol. 2016;100:687–98.
Quigley E. Leaky gut – concept or clinical entity? Curr Opin Gastroenterol. 2016;32:74–9.
Glassner KL, Abraham BP, Quigley EMM. The microbiome and inflammatory bowel disease. J Allergy Clin Immunol. 2020;145:16–27.
Spadoni I, Zagato E, Bertocchi A, Paolinelli R, Hot E, Di Sabatino A, et al. A gut-vascular barrier controls the systemic dissemination of bacteria. Science. 2015;350:830–4.
Bouziat R, Jabri B. IMMUNOLOGY. Breaching the gut-vascular barrier. Science. 2015;350(6262):742–3.
Cheng C, Tan J, Qian W, Zhang L, Hou X. Gut inflammation exacerbates hepatic injury in the high-fat diet induced NAFLD mouse: attention to the gut-vascular barrier dysfunction. Life Sci. 2018;209:157–66.
Huang J, Kelly CP, Bakirtzi K, Villafuerte Gálvez JA, Lyras D, Mileto SJ, et al. Clostridium difficile toxins induce VEGF-A and vascular permeability to promote disease pathogenesis. Nat Microbiol. 2019;4:269–79.
Mouries J, Brescia P, Silvestri A, Spadoni I, Sorribas M, Wiest R, et al. Microbiota-driven gut vascular barrier disruption is a prerequisite for non-alcoholic steatohepatitis development. J Hepatol. 2019;71:1216–28.
Liu P, Bian Y, Fan Y, Zhong J, Liu Z. Protective effect of naringin on in vitro gut-vascular barrier disruption of intestinal microvascular endothelial cells induced by TNF-α. J Agric Food Chem. 2020;68:168–75.
Miyake Y, Yamammoto K. Role of gut microbiota in liver diseases. Hepatol Res. 2013;43:139–46.
Chassaing B, Etienne-Mesmin L, Gewirtz AT. Microbiota-liver axis in hepatic disease. Hepatology. 2014;59(1):328–39.
Seki E, Schnabl B. Role of innate immunity and the microbiota in liver fibrosis: crosstalk between the liver and gut. J Physiol. 2012;590:447–58.
Crispe IN. The liver as a lymphoid organ. Annu Rev Immunol. 2009;27:147–63.
Seo YS, Shah VH. The role of gut liver axis in the pathogenesis of liver cirrhosis and portal hypertension. Clin Mol Hepatol. 2012;18:337–46.
Catala M, Anton A, Portoles MT. Characterization of the simultaneous binding of Escherichia coli endotoxin to Kupffer and endothelial liver cells by flow cytometry. Cytometry. 1999;36:123–30.
Deng M, Scott MJ, Loughran P, Gibson G, Sodhi C, Watkins S, et al. Lipopolysaccharide clearance, bacterial clearance, and systemic inflammatory responses are regulated by cell type-specific functions of TLR4 during sepsis. J Immunol. 2013;190:5152–60.
Hoque R, Vodovotz Y, Mehal W. Therapeutic strategies in inflammasome mediated diseases of the liver. J Hepatol. 2013;58:1047–52.
Patel M, Watson AJM, Rushbrook S. A mechanistic insight into the role of gut microbiota in the pathogenesis of primary sclerosing cholangitis. Gastroenterology. 2019;157:1686–8.
Pastor Rojo O, Lopez San Roman A, Albeniz Arbizu E, et al. Serum lipopolysaccharide–binding protein in endotoxemic patients with inflammatory bowel disease. Inflamm Bowel Dis. 2007;13:269–77.
Terjung B, Spengler U. Atypical p-ANCA in PSC and AIH: a hint toward a “leaky gut”? Clin Rev Allergy Immunol. 2009;36:40–51.
Terjung B, Söhne J, Lechtenberg B, Gottwein J, Muennich M, Herzog V, et al. p-ANCAs in autoimmune liver disorders recognise human beta-tubulin isotype 5 and cross-react with microbial protein FtsZ. Gut. 2010;59:808–16.
Terziroli Beretta-Piccoli B, Mieli-Vergani G, Vergani D, Vierling JM, Adams D, Alpini G, et al. The challenges of primary biliary cholangitis: what is new and what needs to be done. J Autoimmun. 2019;105:102328.
Wang AP, Migita K, Ito M, et al. Hepatic expression of toll–like receptor 4 in primary biliary cirrhosis. J Autoimmun. 2005;25:85–91.
Hopf U, Möller B, Stemerowicz R, et al. Relation between Escherichia coli R (rough)-forms in gut, lipid A in liver, and primary biliary cirrhosis. Lancet. 1989;2:1419–22.
Bogdanos DP, Baum H, Grasso A, et al. Microbial mimics are major targets of crossreactivity with human pyruvate dehydrogenase in primary biliary cirrhosis. J Hepatol. 2004;40:31–9.
Bogdanos DP, Baum H, Okamoto M, et al. Primary biliary cirrhosis is characterized by IgG3 antibodies cross-reactive with the major mitochondrial autoepitope and its Lactobacillus mimic. Hepatology. 2005;42:458–65.
Schwabe RF, Greten TF. Gut microbiome in HCC. J Hepatol. 2020;72:230–8.
Fox JG, Feng Y, Theve EJ, Raczynski AR, Fiala JL, Doernte AL, et al. Gut microbes define liver cancer risk in mice exposed to chemical and viral transgenic heptocarcinogenesis. Gut. 2010;59:88–97.
Yoshimoto S, Loo TM, Atarashi K, Kanda H, Sato S, Oyadomari S, et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature. 2013;499:97–101.
Bindels IB, Porporato P, Dewulf EM, Verrax J, Neyrinck AM, Martin JC, et al. Gut microbiota-derives propionate reduces cancer cell proliferation in the liver. Br J Cancer. 2012;107:1337–44.
Ponziani FR, Bhoori S, Castelli C, Putignani L, Rivoltini L, Del Chierico F, et al. Hepatocellular carcinoma is associated with gut microbiota profile and inflammation in nonalcoholic fatty liver disease. Hepatology. 2019;69:107–20.
Ponziani FR, Nicoletti A, Gasbarrini A, Pompili M. Diagnostic and therapeutic potential of the gut microbiota in patients with early hepatocellular carcinoma. Ther Adv Med Oncol. 2019;11:1758835919848184.
Orci LA, Lacotte S, Delaune V, Slits F, Oldani G, Lazarevic V, et al. Effects of the gut-liver axis on ischaemia-mediated hepatocellular carcinoma recurrence in the mouse liver. J Hepatol. 2018;68:978–85.
Li B, Selmi C, Tang R, Gershwin ME, Ma X. The microbiome and autoimmunity: a paradigm from the gut-liver axis. Cell Mol Immunol. 2018;15:595–609.
Ma HD, Zhao ZB, Ma WT, Liu QZ, Gao CY, Li L, et al. Gut microbiota translocation promotes autoimmune cholangitis. J Autoimmun. 2018;95:47–57.
Denton C, Price A, Friend J, Manithody C, Blomenkamp K, Westrich M, et al. Role of the gut-liver axis in driving parenteral nutrition-associated injury. Children (Basel). 2018;5:pii: E136.
Kummen M, Hov JR. The gut microbial influence on cholestatic liver disease. Liver Int. 2019;39:1186–96.
Wei Y, Li Y, Yan L, Sun C, Miao Q, Wang Q, et al. Alterations of gut microbiome in autoimmune hepatitis. Gut. 2020;69:569–77.
Szabo G. Gut-liver axis beyond the microbiome: how the fungal mycobiome contributes to alcoholic liver disease. Hepatology. 2018;68:2426–8.
Bolte FJ, Rehermann B. Mucosal-invariant T cells in chronic inflammatory liver disease. Semin Liver Dis. 2018;38:60–5.
Marrero I, Maricic I, Feldstein AE, Loomba R, Schnabl B, Rivera-Nieves J, et al. Complex network of NKT cell subsets controls immune homeostasis in liver and gut. Front Immunol. 2018;9:2082.
Biolato M, Manca F, Marrone G, Cefalo C, Racco S, Miggiano GA, et al. Intestinal permeability after Mediterranean diet and low-fat diet in non-alcoholic fatty liver disease. World J Gastroenterol. 2019;25:509–20.
Nakamura K, Kageyama S, Ito T, Hirao H, Kadono K, Aziz A, et al. Antibiotic pretreatment alleviates liver transplant damage in mice and humans. J Clin Invest. 2019;129:3420–34.
Bajaj JS, Hays RA. Manipulation of the gut-liver axis using microbiome restoration therapy in primary sclerosing cholangitis. Am J Gastroenterol. 2019;114:1027–9.
Liu R, Kang JD, Sartor RB, Sikaroodi M, Fagan A, Gavis EA, et al. Neuroinflammation in murine cirrhosis is dependent on the gut microbiome and is attenuated by fecal transplant. Hepatology. 2020;71(2):611–26.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Quigley, E.M.M. (2020). The Microbiota-Gut-Liver Axis: Implications for the Pathophysiology of Liver Disease. In: Gershwin, M.E., M. Vierling, J., Tanaka, A., P. Manns, M. (eds) Liver Immunology . Springer, Cham. https://doi.org/10.1007/978-3-030-51709-0_8
Download citation
DOI: https://doi.org/10.1007/978-3-030-51709-0_8
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-51708-3
Online ISBN: 978-3-030-51709-0
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)