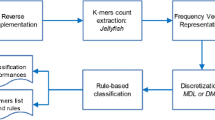
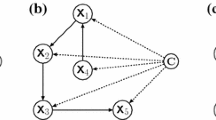
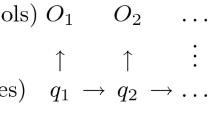Abstract
Most of the related works in Machine Learning (ML) are concerned with Flat Classification, in which an instance is often associated with one class within a small set of classes. However, in some cases, instances have to be assigned to many classes simultaneously, and these classes are arranged in a hierarchical structure. This problem, called Hierarchical Classification (HC), has received special attention in some fields, such as Bioinformatics. In this context, a topic that has gained attention is the classification of Transposable Elements (TEs), which are DNA fragments capable of moving inside the genome of their hosts. In this paper, we propose a novel hierarchical method based on Genetic Algorithms (GAs) that generates HC rules and classifies TEs in many hierarchical levels of its taxonomy. The proposed method is called Hierarchical Classification with a Weighted Genetic Algorithm (HC-WGA), and is based on a Weighted Sum approach to deal with the accuracy-interpretability trade-off, which is a common and still relevant problem in both ML and Bioinformatics. To the best of our knowledge, this is the first HC method to use such an approach. Experiments with two popular TEs datasets showed that our method achieves competitive results with most of the state-of-the-art HC methods, with the advantage of presenting an interpretable model.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
Notes
- 1.
Available at https://github.com/geantrindade/HC-WGA (20 June 2019).
- 2.
Available at http://pgsb.helmholtz-muenchen.de/plant/ (20 June 2019).
- 3.
Available at http://girinst.org/repbase/ (20 June 2019).
- 4.
Available at https://github.com/geantrindade/TEsHierarchicalDatasets (20 June 2019).
References
Bandaru, S., Ng, A.H., Deb, K.: Data mining methods for knowledge discovery in multi-objective optimization: part a-survey. Expert Syst. Appl. 70, 139–159 (2017)
Costa, E.P., Lorena, A.C., Carvalho, A.C.P.L.F., Freitas, A.A., Holden, N.: Comparing several approaches for hierarchical classification of proteins with decision trees. In: Sagot, M.-F., Walter, M.E.M.T. (eds.) BSB 2007. LNCS, vol. 4643, pp. 126–137. Springer, Heidelberg (2007). https://doi.org/10.1007/978-3-540-73731-5_12
Finnegan, D.J.: Eukaryotic transposable elements and genome evolution. Trends Genet. 5, 103–107 (1989)
Freitas, A.A.: A critical review of multi-objective optimization in data mining: a position paper. SIGKDD Explor. Newsl. 6(2), 77–86 (2004)
Hollander, M., Wolfe, D.A., Chicken, E.: Nonparametric Statistical Methods. Wiley, New York (2013)
Jurka, J., Kapitonov, V.V., Pavlicek, A., Klonowski, P., Kohany, O., Walichiewicz, J.: Repbase update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 110(1–4), 462–467 (2005)
Kiritchenko, S., Matwin, S., Nock, R., Famili, A.F.: Learning and evaluation in the presence of class hierarchies: application to text categorization. In: Lamontagne, L., Marchand, M. (eds.) AI 2006. LNCS (LNAI), vol. 4013, pp. 395–406. Springer, Heidelberg (2006). https://doi.org/10.1007/11766247_34
Lipton, Z.C.: The mythos of model interpretability. ar**v preprint ar**v:1606.03490 (2016)
McClintock, B.: The Significance of Responses of the Genome to Challenge. World Scientific Pub. Co., Singapore (1993)
Nakano, F.K., Mastelini, S.M., Barbon, S., Cerri, R.: Improving hierarchical classification of transposable elements using deep neural networks. In: 2018 International Joint Conference on Neural Networks (IJCNN), pp. 1–8. IEEE (2018)
Nakano, F.K., Pinto, W.J., Pappa, G.L., Cerri, R.: Top-down strategies for hierarchical classification of transposable elements with neural networks. In: 2017 International Joint Conference on Neural Networks (IJCNN), pp. 2539–2546. IEEE (2017)
Nussbaumer, T., et al.: MIPS PlantsDB: a database framework for comparative plant genome research. Nucleic Acids Res. 41(D1), D1144–D1151 (2012)
Pereira, G.T., Cerri, R.: Hierarchical and non-hierarchical classification of transposable elements with a genetic algorithm. J. Inf. Data Manage. 9(1), 163–178 (2018)
Pereira, G.T., Santos, B.Z., Cerri, R.: A genetic algorithm for transposable elements hierarchical classification rule induction. In: 2018 IEEE Congress on Evolutionary Computation (CEC), pp. 1–8. IEEE (2018)
Poursabzi-Sangdeh, F., Goldstein, D.G., Hofman, J.M., Vaughan, J.W., Wallach, H.: Manipulating and measuring model interpretability. ar**v preprint ar**v:1802.07810 (2018)
Santos, B.Z., Pereira, G.T., Nakano, F.K., Cerri, R.: Strategies for selection of positive and negative instances in the hierarchical classification of transposable elements. In: 2018 7th Brazilian Conference on Intelligent Systems (BRACIS), pp. 420–425. IEEE (2018)
Silla, C.N., Freitas, A.A.: A survey of hierarchical classification across different application domains. Data Min. Knowl. Disc. 22(1–2), 31–72 (2011)
Vens, C., Struyf, J., Schietgat, L., Džeroski, S., Blockeel, H.: Decision trees for hierarchical multilabel classification. Mach. Learn. 73(2), 185–214 (2008)
Wicker, T., et al.: A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 8(12), 973–982 (2007)
Acknowledgment
This study was financed by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brazil (CAPES) - Finance Code 001, as well as by the Sao Paulo Research Foundation (FAPESP), grants 2015/14300-1 and 2016/50457-5.
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Pereira, G.T., Gabriel, P.H.R., Cerri, R. (2019). Hierarchical Classification of Transposable Elements with a Weighted Genetic Algorithm. In: Moura Oliveira, P., Novais, P., Reis, L. (eds) Progress in Artificial Intelligence. EPIA 2019. Lecture Notes in Computer Science(), vol 11804. Springer, Cham. https://doi.org/10.1007/978-3-030-30241-2_61
Download citation
DOI: https://doi.org/10.1007/978-3-030-30241-2_61
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-30240-5
Online ISBN: 978-3-030-30241-2
eBook Packages: Computer ScienceComputer Science (R0)