Abstract
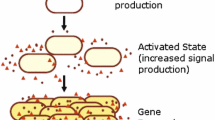
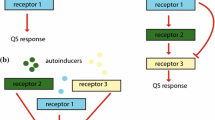
Certain species of bacteria are capable of communicating through a mechanism called Quorum Sensing (QS) wherein they release and sense signaling molecules, called autoinducers, to and from the environment. Despite stochastic fluctuations, bacteria gradually achieve coordinated gene expression through QS, which in turn, help them better adapt to environmental adversities. Existing sequential approaches for modeling information exchange via QS for large cell populations are time and computational resource intensive, because the advancement in simulation time becomes significantly slower with the increase in molecular concentration. This paper presents a scalable parallel framework for modeling multicellular communication. Simulations show that our framework accurately models the molecular concentration dynamics of QS system, yielding better speed-up and CPU utilization than the existing sequential model that uses the exact Gillespie algorithm. We also discuss how our framework accommodates evolving population due to cell birth, death and heterogeneity due to noise. Furthermore, we analyze the performance of our framework vis-á-vis the effects of its data sampling interval and Gillespie computation time. Finally, we validate the scalability of the proposed framework by modeling population size up to 2000 bacterial cells.
S. Roy and M. A. Islam—Primary co-authors.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Miller, M.B., Bassler, B.L.: Quorum sensing in bacteria. Ann. Rev. Microbiol. 55(1), 165–199 (2001)
Weber, M., Buceta, J.: Dynamics of the quorum sensing switch: stochastic and non-stationary effects. BMC Syst. Biol. 7, 6 (2013)
Boada, Y., Vignoni, A., Pico, J.: Promoter and transcription factor dynamics tune protein mean and noise strength in a quorum sensing-based feedback synthetic circuit. bioRxiv, p. 106229 (2017)
Bassler, B.: Manipulating quorum sensing to control bacterial pathogenicity. FASEB J. 29(1 Suppl.), 88-1 (2015)
Kang, S., Kahan, S., McDermott, J., Flann, N., Shmulevich, I.: Biocellion: accelerating computer simulation of multicellular biological system models. Bioinformatics 30(21), 3101–3108 (2014)
Tian, T., Burrage, K.: Parallel implementation of stochastic simulation for large-scale cellular processes. In: Proceedings of the Eighth International Conference on High-Performance Computing in Asia-Pacific Region, pp. 6–pp. IEEE (2005)
Li, H., Cao, Y., Petzold, L.R., Gillespie, D.T.: Algorithms and software for stochastic simulation of biochemical reacting systems. Biotechnol. Progress 24(1), 56–61 (2008)
Komarov, I., D’Souza, R.M.: Accelerating the gillespie exact stochastic simulation algorithm using hybrid parallel execution on graphics processing units. PloS One 7(11), e46693 (2012)
Harvey, D.G., Fletcher, A.G., Osborne, J.M., Pitt-Francis, J.: A parallel implementation of an off-lattice individual-based model of multicellular populations. Comput. Phys. Commun. 192, 130–137 (2015)
Kouskoumvekakis, E., Soudris, D., Manolakos, E.S.: Many-core CPUs can deliver scalable performance to stochastic simulations of large-scale biochemical reaction networks. In: 2015 International Conference on High Performance Computing & Simulation (HPCS). IEEE (2015)
Vanneschi, M.: The programming model of assist, an environment for parallel and distributed portable applications. Parallel Comput. 28(12), 1709–1732 (2002)
Dematté, L., Mazza, T.: On parallel stochastic simulation of diffusive systems. In: Heiner, M., Uhrmacher, A.M. (eds.) CMSB 2008. LNCS (LNAI), vol. 5307, pp. 191–210. Springer, Heidelberg (2008). https://doi.org/10.1007/978-3-540-88562-7_16
Islam, M.A., Roy, S., Das, S., Barua, D.: Multicellular models bridging intracellular signaling and gene transcription to population dynamics. Processes 6(11), 217 (2018)
Boada, Y., Vignoni, A., Navarro, J.L., Picó, J.: Improvement of a CLE stochastic simulation of gene synthetic network with quorum sensing and feedback in a cell population. In: 2015 European Control Conference (ECC), pp. 2274–2279. IEEE (2015)
Oudkerk, R., Noller, J.: Multiprocessing process-based threading interface. https://docs.python.org/2/library/multiprocessing.html
Boada, Y., Vignoni, A., Picó, J.: Engineered control of genetic variability reveals interplay among quorum sensing, feedback regulation, and biochemical noise. ACS Synth. Biol. 6(10), 1903–1912 (2017)
Rodola, G.: Psutil: cross-platform lib for process and system monitoring in python. https://pypi.org/project/psutil/
Acknowledgement
The research presented in this work is supported by the National Science Foundation CBET-CDS&E grant 1609642.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 ICST Institute for Computer Sciences, Social Informatics and Telecommunications Engineering
About this paper
Cite this paper
Roy, S., Islam, M.A., Barua, D., Das, S.K. (2019). A Scalable Parallel Framework for Multicellular Communication in Bacterial Quorum Sensing. In: Compagnoni, A., Casey, W., Cai, Y., Mishra, B. (eds) Bio-inspired Information and Communication Technologies. BICT 2019. Lecture Notes of the Institute for Computer Sciences, Social Informatics and Telecommunications Engineering, vol 289. Springer, Cham. https://doi.org/10.1007/978-3-030-24202-2_14
Download citation
DOI: https://doi.org/10.1007/978-3-030-24202-2_14
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-24201-5
Online ISBN: 978-3-030-24202-2
eBook Packages: Computer ScienceComputer Science (R0)