Abstract
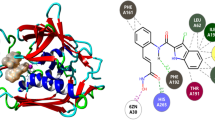
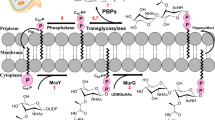
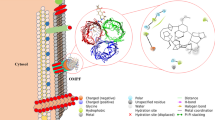
An increase in the number of antibiotic-resistant bacterial pathogens, in recent times, has posed a great challenge for treating the affected patients. This has paved the way for the development and design of antibiotics against the previously less explored newer targets. Among these, peptidoglycan (PG) biosynthesis serves as a promising target for the design and development of novel drugs. The peptidoglycan cell wall synthesis in bacteria is essential for its viability. The enzyme class, Mur ligases, plays a key role in PG biosynthesis. Therefore, compounds with the ability to inhibit these enzymes (Mur ligase) can serve as potential candidates for develo** small modulators. The enzyme, UDP-N-acetyl pyruvyl-glucosamine reductase (MurB), is essential for PG biosynthesis, a crucial part of the bacterial cell wall. The development of novel drugs to treat infections may thus focus on inhibiting MurB function. Understanding the mechanism of action of Mur B is central to develo** efficient inhibitors. For the treatment of S. typhi infections, it is also critical to find therapeutic drugs that specifically target MurB. The enzyme Mur B from Salmonella enterica serovar Typhi (stMurB) was expressed and purified for biophysical characterization to gauge the molecular interactions and estimate thermodynamic stability, for determining attributes for possible therapeutic intervention. The thermal melting profile of MurB was monitored by circular dichroism (CD) and validated by performing differential scanning calorimetry (DSC). An in silico virtual screening of various natural inhibitors was conducted with modelled stMurB structure. The three top hits (quercetin, berberine, and scopoletin) obtained from in silico screening were validated for complex stability through molecular dynamics (MD) simulation. Further, fluorescence binding studies were undertaken for the selected natural inhibitors with stMurB alone and with its NADPH-bound form. The natural inhibitors, scopoletin and berberine, displayed lesser binding to stMurB compared to quercetin. Also, a stronger binding affinity was exhibited between quercetin and stMurB compared to NADPH and stMurB. Based on the above two findings, quercetin can be developed as an inhibitor of stMurB enzyme.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Bugg TD, Braddick D, Dowson CG, Roper DI (2011) Bacterial cell wall assembly: still an attractive antibacterial target. Trends Biotechnol 29(4):167–173. https://doi.org/10.1016/j.tibtech.2010.12.006
Hrast M, Sosic I, Sink R, Gobec S (2014) Inhibitors of the peptidoglycan biosynthesis enzymes MurA-F. Bioorg Chem 55:2–15. https://doi.org/10.1016/j.bioorg.2014.03.008
Gautam A, Vyas R, Tewari R (2011) Peptidoglycan biosynthesis machinery: a rich source of drug targets. Crit Rev Biotechnol 31(4):295–336. https://doi.org/10.3109/07388551.2010.525498
Silhavy TJ, Kahne D, Walker S (2010) The bacterial cell envelope. Cold Spring Harb Perspect Biol 2(5):a000414. https://doi.org/10.1101/cshperspect.a000414
El Zoeiby A, Sanschagrin F, Levesque RC (2003) Structure and function of the Mur enzymes: development of novel inhibitors. Mol Microbiol 47(1):1–12. https://doi.org/10.1046/j.1365-2958.2003.03289.x
Bratkovic T, Lunder M, Urleb U, Strukelj B (2008) Peptide inhibitors of MurD and MurE, essential enzymes of bacterial cell wall biosynthesis. J Basic Microbiol 48(3):202–206. https://doi.org/10.1002/jobm.200700133
Perdih A, Kovac A, Wolber G, Blanot D, Gobec S, Solmajer T (2009) Discovery of novel benzene 1,3-dicarboxylic acid inhibitors of bacterial MurD and MurE ligases by structure-based virtual screening approach. Bioorg Med Chem Lett 19(10):2668–2673. https://doi.org/10.1016/j.bmcl.2009.03.141
Horton JR, Bostock JM, Chopra I, Hesse L, Phillips SE, Adams DJ, Johnson AP, Fishwick CW (2003) Macrocyclic inhibitors of the bacterial cell wall biosynthesis enzyme MurD. Bioorg Med Chem Lett 13(9):1557–1560
Tomasic T, Sink R, Zidar N, Fic A, Contreras-Martel C, Dessen A, Patin D, Blanot D, Muller-Premru M, Gobec S, Zega A, Kikelj D, Masic LP (2012) Dual inhibitor of MurD and MurE ligases from Escherichia coli and Staphylococcus aureus. ACS Med Chem Lett 3(8):626–630. https://doi.org/10.1021/ml300047h
Zivec M, Turk S, Blanot D, Gobec S (2011) Design and synthesis of new peptidomimetics as potential inhibitors of MurE. Acta Chim Slov 58(1):95–109
Reynaud E (2010) Protein misfolding and degenerative diseases. Nat Educ 3(9):28
Kumar GR, Sharma A, Kumari M, Jagannadham MV, Debnath M (2011) Equilibrium unfolding of A. niger RNase: pH dependence of chemical and thermal denaturation. Eur Biophys J 40(8):923–935. https://doi.org/10.1007/s00249-011-0708-1
Alam P, Abdelhameed AS, Rajpoot RK, Khan RH (2016) Interplay of multiple interaction forces: binding of tyrosine kinase inhibitor nintedanib with human serum albumin. J Photochem Photobiol B 157:70–76. https://doi.org/10.1016/j.jphotobiol.2016.02.009
Nick Pace C, Scholtz JM, Grimsley GR (2014) Forces stabilizing proteins. FEBS Lett 588(14):2177–2184. https://doi.org/10.1016/j.febslet.2014.05.006
Kim PS, Baldwin RL (1990) Intermediates in the folding reactions of small proteins. Annu Rev Biochem 59:631–660. https://doi.org/10.1146/annurev.bi.59.070190.003215
Matthews CR (1993) Pathways of protein folding. Annu Rev Biochem 62:653–683. https://doi.org/10.1146/annurev.bi.62.070193.003253
Clark AC (2008) Protein folding: are we there yet? Arch Biochem Biophys 469(1):1–3. https://doi.org/10.1016/j.abb.2007.10.007
Ubaid-Ullah S, Haque MA, Zaidi S, Hassan MI, Islam A, Batra JK, Singh TP, Ahmad F (2014) Effect of sequential deletion of extra N-terminal residues on the structure and stability of yeast iso-1-cytochrome-c. J Biomol Struct Dyn 32(12):2005–2016. https://doi.org/10.1080/07391102.2013.848826
Haque MA, Singh M, Tripathi MK, Ethayathulla AS, Kaur P (2023) Identification of natural small molecule modulators of MurB from Salmonella enterica serovar Typhi Ty2 strain using computational and biophysical approaches. Proteins 91(3):363–379. https://doi.org/10.1002/prot.26435
Miles AJ, Ramalli SG, Wallace BA (2022) DichroWeb, a website for calculating protein secondary structure from circular dichroism spectroscopic data. Protein Sci 31(1):37–46. https://doi.org/10.1002/pro.4153
Whitmore L, Wallace BA (2008) Protein secondary structure analyses from circular dichroism spectroscopy: methods and reference databases. Biopolymers 89(5):392–400. https://doi.org/10.1002/bip.20853
Ahmad F, Bigelow CC (1982) Estimation of the free energy of stabilization of ribonuclease A, lysozyme, alpha-lactalbumin, and myoglobin. J Biol Chem 257(21):12935–12938
Shirley BA (1995) Urea and guanidine hydrochloride denaturation curves. Methods Mol Biol 40:177–190. https://doi.org/10.1385/0-89603-301-5:177
Benson TE, Walsh CT, Hogle JM (1997) X-ray crystal structures of the S229A mutant and wild-type MurB in the presence of the substrate enolpyruvyl-UDP-N-acetylglucosamine at 1.8-A resolution. Biochemistry 36(4):806–811. https://doi.org/10.1021/bi962221g
Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Gallo Cassarino T, Bertoni M, Bordoli L, Schwede T (2014) SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 42(Web server issue):W252–W258. https://doi.org/10.1093/nar/gku340
Laskowski RA, Moss DS, Thornton JM (1993) Main-chain bond lengths and bond angles in protein structures. J Mol Biol 231(4):1049–1067. https://doi.org/10.1006/jmbi.1993.1351
Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM (1996) AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR 8(4):477–486. https://doi.org/10.1007/BF00228148
Luthy R, Bowie JU, Eisenberg D (1992) Assessment of protein models with three-dimensional profiles. Nature 356(6364):83–85. https://doi.org/10.1038/356083a0
Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC, Mainz DT (2006) Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J Med Chem 49(21):6177–6196. https://doi.org/10.1021/jm051256o
2023-1 SR (2021) Maestro, Schrödinger. LLC, New York
Bowers KJ, Chow E, Xu H, Dror RO, Eastwood MP, Gregersen BA, Klepeis JL, Kolossvary I, Moraes MA, Sacerdoti FD, Salmon JK, Shan Y, Shaw DE (2006) Scalable algorithms for molecular dynamics simulations on commodity clusters. In: Proceedings of the ACM/IEEE conference on supercomputing (SC06), Tampa, Florida, November 11–17, 2006
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23(21):2947–2948. https://doi.org/10.1093/bioinformatics/btm404
Notredame C, Higgins DG, Heringa J (2000) T-coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol 302(1):205–217
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30(4):772–780. https://doi.org/10.1093/molbev/mst010
Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, Lepore R, Schwede T (2018) SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46(W1):W296–W303. https://doi.org/10.1093/nar/gky427
Baek M, DiMaio F, Anishchenko I, Dauparas J, Ovchinnikov S, Lee GR, Wang J, Cong Q, Kinch LN, Schaeffer RD, Millan C, Park H, Adams C, Glassman CR, DeGiovanni A, Pereira JH, Rodrigues AV, van Dijk AA, Ebrecht AC, Opperman DJ, Sagmeister T, Buhlheller C, Pavkov-Keller T, Rathinaswamy MK, Dalwadi U, Yip CK, Burke JE, Garcia KC, Grishin NV, Adams PD, Read RJ, Baker D (2021) Accurate prediction of protein structures and interactions using a three-track neural network. Science 373(6557):871–876. https://doi.org/10.1126/science.abj8754
Jo S, Kim T, Iyer VG, Im W (2008) CHARMM-GUI: a web-based graphical user interface for CHARMM. J Comput Chem 29(11):1859–1865. https://doi.org/10.1002/jcc.20945
Brooks BR, Brooks CL 3rd, Mackerell AD Jr, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M (2009) CHARMM: the biomolecular simulation program. J Comput Chem 30(10):1545–1614. https://doi.org/10.1002/jcc.21287
Lee J, Cheng X, Swails JM, Yeom MS, Eastman PK, Lemkul JA, Wei S, Buckner J, Jeong JC, Qi Y, Jo S, Pande VS, Case DA, Brooks CL 3rd, MacKerell AD Jr, Klauda JB, Im W (2016) CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J Chem Theory Comput 12(1):405–413. https://doi.org/10.1021/acs.jctc.5b00935
Lee J, Hitzenberger M, Rieger M, Kern NR, Zacharias M, Im W (2020) CHARMM-GUI supports the Amber force fields. J Chem Phys 153(3):035103. https://doi.org/10.1063/5.0012280
2023-1 SR (2021) LigPrep, Schrödinger. LLC, New York
Greenwood JR, Calkins D, Sullivan AP, Shelley JC (2010) Towards the comprehensive, rapid, and accurate prediction of the favorable tautomeric states of drug-like molecules in aqueous solution. J Comput Aided Mol Des 24(6-7):591–604. https://doi.org/10.1007/s10822-010-9349-1
Harder E, Damm W, Maple J, Wu C, Reboul M, **ang JY, Wang L, Lupyan D, Dahlgren MK, Knight JL, Kaus JW, Cerutti DS, Krilov G, Jorgensen WL, Abel R, Friesner RA (2016) OPLS3: a force field providing broad coverage of drug-like small molecules and proteins. J Chem Theory Comput 12(1):281–296. https://doi.org/10.1021/acs.jctc.5b00864
Halgren TA (2009) Identifying and characterizing binding sites and assessing druggability. J Chem Inf Model 49(2):377–389. https://doi.org/10.1021/ci800324m
Zhu K, Day T, Warshaviak D, Murrett C, Friesner R, Pearlman D (2014) Antibody structure determination using a combination of homology modeling, energy-based refinement, and loop prediction. Proteins 82(8):1646–1655. https://doi.org/10.1002/prot.24551
Salam NK, Adzhigirey M, Sherman W, Pearlman DA (2014) Structure-based approach to the prediction of disulfide bonds in proteins. Protein Eng Des Sel 27(10):365–374. https://doi.org/10.1093/protein/gzu017
Beard H, Cholleti A, Pearlman D, Sherman W, Loving KA (2013) Applying physics-based scoring to calculate free energies of binding for single amino acid mutations in protein-protein complexes. PLoS One 8(12):e82849. https://doi.org/10.1371/journal.pone.0082849
Sastry GM, Adzhigirey M, Day T, Annabhimoju R, Sherman W (2013) Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des 27(3):221–234. https://doi.org/10.1007/s10822-013-9644-8
Berendsen HJC, van der Spoel D, van Drunen RV (1995) GROMACS: a message-passing parallel molecular dynamics implementation. Comput Phys Commun 91(1–3):43–56. https://doi.org/10.1016/0010-4655(95)00042-E
Pronk S, Pall S, Schulz R, Larsson P, Bjelkmar P, Apostolov R, Shirts MR, Smith JC, Kasson PM, van der Spoel D, Hess B, Lindahl E (2013) GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 29(7):845–854. https://doi.org/10.1093/bioinformatics/btt055
Hess B, Kutzner C, van der Spoel D, Lindahl E (2008) GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput 4(3):435–447. https://doi.org/10.1021/ct700301q
Acknowledgments
MK and MAH thank the Indian Council of Medical Research, New Delhi for the grant of fellowship.
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2024 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Kumar, M., Haque, M.A., Kaur, P. (2024). Computational and Biophysical Approaches to Identify Cell Wall-Associated Modulators in Salmonella enterica serovar Typhi. In: Ton-That, H. (eds) The Bacterial Cell Wall. Methods in Molecular Biology, vol 2727. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-3491-2_4
Download citation
DOI: https://doi.org/10.1007/978-1-0716-3491-2_4
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-3490-5
Online ISBN: 978-1-0716-3491-2
eBook Packages: Springer Protocols