Abstract
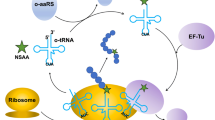
The incorporation of nonstandard amino acids (nsAAs) within protein sequences has broadened the chemical functionalities available for use in the study, prevention, or treatment of disease. The ability to genetically encode the introduction of nsAAs at precise sites of target recombinant proteins has enabled numerous applications such as bioorthogonal conjugation, thrombin inhibition, intrinsic biological containment of live organisms, and immunochemical termination of self-tolerance. Genetic systems that perform critical steps in enabling nsAA incorporation are known as orthogonal translation systems or orthogonal aminoacyl-tRNA synthetase/tRNA pairs. In Escherichia coli, several of these have been designed to accept novel nsAAs. Certain endogenous proteins, codon context, and standard amino acid concentrations can affect the yield of recombinant protein, the rate of nsAA incorporation within off-target proteins, and the rate of misincorporation due to near-cognate suppression or misacylation of orthogonal tRNA with standard amino acids. As a result, a significant body of work has been performed in engineering the E. coli genome to alleviate these issues. Here, we describe common methods applicable to nsAA incorporation within proteins in E. coli for sufficient purity and characterization for downstream therapeutic applications.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Sletten EM, Bertozzi CR (2009) Bioorthogonal chemistry: fishing for selectivity in a sea of functionality. Angew Chem Int Ed 48:6974–6998
Kolb HC, Finn MG, Sharpless KB (2001) Click chemistry: diverse chemical function from a few good reactions. Angew Chem Int Ed 40:2004–2021
Cho H, Daniel T, Buechler YJ et al (2011) Optimized clinical performance of growth hormone with an expanded genetic code. Proc Nat Acad Sci 108:9060–9065
Hallam TJ, Wold E, Wahl A et al (2015) Antibody conjugates with unnatural amino acids. Mol Pharmaceutics 12(6):1848–1862
Kim CH, Axup JY, Dubrovska A et al (2012) Synthesis of bispecific antibodies using genetically encoded unnatural amino acids. J Am Chem Soc 134:9918–9921
Agarwal P, Bertozzi CR (2015) Site-specific antibody-drug conjugates: the nexus of bioorthogonal chemistry, protein engineering, and drug development. Bioconjugate Chem 26:176–192
Tian F, Lu Y, Manibusan A et al (2014) A general approach to site-specific antibody drug conjugates. Proc Natl Acad Sci U S A 111:1766–1771
Roy G, Reier J, Garcia A et al (2020) Development of a high yielding expression platform for the introduction of non-natural amino acids in protein sequences. MAbs 12:1684749
BrickBio, https://www.brickbio.com/
Groff D, Carlos NA, Chen R et al (2022) Development of an E coli strain for cell-free ADC manufacturing. Biotechnol Bioeng 119:162–175
Milla ME, Ptacin JL, Ma L et al (2019) THOR-707, a novel not-alpha IL-2, promotes all key immune system anti-tumoral actions of IL-2 without eliciting vascular leak syndrome (VLS). Ann Oncol 30:v501
Liu CC, Schultz PG (2006) Recombinant expression of selectively sulfated proteins in Escherichia coli. Nat Biotechnol 24:1436–1440
Liu CC, Choe H, Farzan M, Smider VV, Schultz PG (2009) Mutagenesis and evolution of sulfated antibodies using an expanded genetic code. Biochemistry 48(37):8891–8898
Chen S, Maini R, Bai X et al (2017) Incorporation of phosphorylated tyrosine into proteins: in vitro translation and study of phosphorylated IκB-α and its interaction with NF-κB. J Am Chem Soc 139:14098–14108
Grünewald J, Tsao ML, Perera R et al (2008) Immunochemical termination of self-tolerance. Proc Natl Acad Sci U S A 105:11276–11280
Gauba V, Grunewald J, Gorney V et al (2011) Loss of CD4 T-cell-dependent tolerance to proteins with modified amino acids. Proc Nat Acad Sci 108:12821–12826
Hemphill J, Borchardt EK, Brown K et al (2015) Optical control of CRISPR/Cas9 gene editing. J Am Chem Soc 137:5642–5645
Jones CM, Robkis DM, Blizzard RJ et al (2021) Genetic encoding of a highly photostable, long lifetime fluorescent amino acid for imaging in mammalian cells. Proc Nat Acad Sci 12:11955–11964
Thyer R, Shroff R, Klein DR et al (2018) Custom selenoprotein production enabled by laboratory evolution of recoded bacterial strains. Nat Biotechnol 36:624–631
Yuan Z, Wang N, Kang G et al (2017) Controlling multicycle replication of live-attenuated HIV-1 using an unnatural genetic switch. ACS Synth Biol 6:721–731
Wang N, Li Y, Niu W et al (2014) Construction of a live-attenuated HIV-1 vaccine through genetic code expansion. Angew Chem Int Ed Engl 53:4867–4871
Mandell DJ, Lajoie MJ, Mee MT et al (2015) Biocontainment of genetically modified organisms by synthetic protein design. Nature 518:55–60
Kunjapur AM, Napolitano MG, Hysolli E et al (2021) Synthetic auxotrophy remains stable after continuous evolution and in coculture with mammalian cells. Science 7:eabf5851
Johnson JA, Lu YY, Van DJA et al (2010) Residue-specific incorporation of non-canonical amino acids into proteins: recent developments and applications. Curr Opin Chem Biol 14:774–780
Singh-Blom A, Hughes RA, Ellington AD (2013) Residue-specific incorporation of unnatural amino acids into proteins in vitro and in vivo. In: Methods in molecular biology. Humana Press, Totowa, pp 93–114
Anderson JC, Wu N, Santoro SW et al (2004) An expanded genetic code with a functional quadruplet codon. Proc Natl Acad Sci USA 101:7566–7571
Magliery TJ, Anderson JC, Schultz PG (2001) Expanding the genetic code: selection of efficient suppressors of four-base codons and identification of “shifty” four-base codons with a library approach in Escherichia coli. J Mol Biol 307:755–769
DeBenedictis EA, Carver GD, Chung CZ et al (2021) Multiplex suppression of four quadruplet codons via tRNA directed evolution. Nat Commun 12:1–13
Neumann H, Wang K, Davis L et al (2010) Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome. Nature 464:441–444
Schmied WH, Tnimov Z, Uttamapinant C et al (2018) Controlling orthogonal ribosome subunit interactions enables evolution of new function. Nature 564:444–448
Malyshev DA, Dhami K, Lavergne T et al (2014) A semi-synthetic organism with an expanded genetic alphabet. Nature 509:385–388
Zhang Y, Ptacin JL, Fischer EC et al (2017) A semi-synthetic organism that stores and retrieves increased genetic information. Nature 551:644–647
Fischer EC, Hashimoto K, Zhang Y et al (2020) New codons for efficient production of unnatural proteins in a semisynthetic organism. Nat Chem Biol 16(5):570–576
Wan W, Tharp JM, Liu WR (2014) Pyrrolysyl-tRNA synthetase: an ordinary enzyme but an outstanding genetic code expansion tool. Biochim Biophys Acta BBA Proteins Proteom 1844(6):1059–1070
Dumas A, Lercher L, Spicer CD et al (2015) Designing logical codon reassignment-expanding the chemistry in biology. Chem Sci 6:50–69
Italia JS, Addy PS, Wrobel CJJ et al (2017) An orthogonalized platform for genetic code expansion in both bacteria and eukaryotes. Nat Chem Biol 13:446–450
Zhao H, Ding W, Zang J et al (2021) Directed-evolution of translation system for efficient unnatural amino acids incorporation and generalizable synthetic auxotroph construction. Nat Commun 12:1–12
Young TS, Ahmad I, Yin JA et al (2010) An enhanced system for unnatural amino acid mutagenesis in E. coli. J Mol Biol 395:361–374
Chatterjee A, Sun SB, Furman JL et al (2013) A versatile platform for single- and multiple-unnatural amino acid mutagenesis in Escherichia coli. Biochemistry 52:1828–1837
Lajoie MJ, Rovner AJ, Goodman DB et al (2013) Genomically recoded organisms expand biological functions. Science 342(1979):357–360
Wang L, Schultz PG (2001) A general approach for the generation of orthogonal tRNAs. Chem Biol 8:883–890
Miller SM, Wang T, Liu DR (2020) Phage-assisted continuous and non-continuous evolution. Nat Protoc 15:4101–4127
Kunjapur AM, Stork DA, Kuru E et al (2018) Engineering posttranslational proofreading to discriminate nonstandard amino acids. Proc Natl Acad Sci USA 115:619–624
Chin JW, Santoro SW, Martin AB et al (2002) Addition of p-azido-L-phenylalanine to the genetic code of Escherichia coli. J Am Chem Soc 124:9026–9027
Miyake-Stoner SJ, Refakis CA, Hammill JT et al (2010) Generating permissive site-specific unnatural aminoacyl-tRNA synthetases. Biochemistry 49:1667–1677
Young DD, Young TS, Jahnz M et al (2011) An evolved aminoacyl-tRNA synthetase with atypical polysubstrate specificity. Biochemistry 50:1894–1900
Hohl A, Karan R, Akal A et al (2019) Engineering a polyspecific pyrrolysyl-tRNA synthetase by a high-throughput FACS screen. Sci Rep 9:1–9
Young DD, Jockush S, Turro NJ et al (2011) Synthetase polyspecificity as a tool to modulate protein function. Bioorg Med Chem Lett 21:7502
Ng PC, Henikoff S (2003) SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res 31:3812–3814
Choi Y, Chan AP (2015) PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 31:2745–2747
Gallagher RR, Li Z, Lewis AO et al (2014) Rapid editing and evolution of bacterial genomes using libraries of synthetic DNA. Nat Protoc 9:2301–2316
Wannier TM, Nyerges A, Kuchwara HM et al (2020) Improved bacterial recombineering by parallelized protein discovery. Proc Natl Acad Sci USA 117:13689–13698
Farasat I, Kushwaha M, Collens J et al (2014) Efficient search, map**, and optimization of multi-protein genetic systems in diverse bacteria. Mol Syst Biol 10:731
Chen RH, Huang C-J, Newton BS et al (2009) Factors affecting endotoxin removal from recombinant therapeutic proteins by anion exchange chromatography. Protein Expr Purif 64:76–81
Wang L, Brock A, Herberich B et al (2001) Expanding the genetic code of Escherichia coli. Science 292(1979):498–500
Lee BS, Kim S, Ko BJ et al (2017) An efficient system for incorporation of unnatural amino acids in response to the four-base codon AGGA in Escherichia coli. Biochim Biophys Acta BBA Gen Subj 1861:3016–3023
Venkat S, Sturges J, Stahman A et al (2018) Genetically incorporating two distinct post-translational modifications into one protein simultaneously. ACS Synth Biol 7:689–695
Wan W, Huang Y, Wang Z et al (2010) A facile system for genetic incorporation of two different noncanonical amino acids into one protein in Escherichia coli. Angew Chem Int Ed 49:3211–3214
Blight SK, Larue RC, Mahapatra A et al (2004) Direct charging of tRNACUA with pyrrolysine in vitro and in vivo. Nature 431:333–335
Mukai T, Yamaguchi A, Ohtake K et al (2015) Reassignment of a rare sense codon to a non-canonical amino acid in Escherichia coli. Nucleic Acids Res 43:8111–8122
Ding W, Zhao H, Chen Y et al (2020) Chimeric design of pyrrolysyl-tRNA synthetase/tRNA pairs and canonical synthetase/tRNA pairs for genetic code expansion. Nat Commun 11:1–13
Kuznetsov G, Goodman DB, Filsinger GT et al (2017) Optimizing complex phenotypes through model-guided multiplex genome engineering. Genome Biol 18:1–12
Acknowledgments
This work was supported by the National Science Foundation (NSF CBET #2032243).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2024 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Butler, N.D., Kunjapur, A.M. (2024). Selective and Site-Specific Incorporation of Nonstandard Amino Acids Within Proteins for Therapeutic Applications. In: Sullivan, M.O., Chackerian, B., Chen, W. (eds) Therapeutic Proteins. Methods in Molecular Biology, vol 2720. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-3469-1_3
Download citation
DOI: https://doi.org/10.1007/978-1-0716-3469-1_3
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-3468-4
Online ISBN: 978-1-0716-3469-1
eBook Packages: Springer Protocols