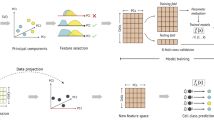
Abstract
Peripheral blood mononuclear cells (PBMC) are mixed subpopulations of blood cells composed of five cell types. PBMC are widely used in the study of the immune system, infectious diseases, cancer, and vaccine development. Single-cell transcriptomics (SCT) allows the labeling of cell types by gene expression patterns from biological samples. Classifying cells into cell types and states is essential for single-cell analyses, especially in the classification of diseases and the assessment of therapeutic interventions, and for many secondary analyses. Most of the classification of cell types from SCT data use unsupervised clustering or a combination of unsupervised and supervised methods including manual correction. In this chapter, we describe a protocol that uses supervised machine learning (ML) methods with SCT data for the classification of PBMC cell types in samples representing pathological states. This protocol has three parts: (1) data preprocessing, (2) labeling of reference PBMC SCT datasets and training supervised ML models, and (3) labeling new PBMC datasets from disease samples. This protocol enables building classification models that are of high accuracy and efficiency. Our example focuses on 10× Genomics technology but applies to datasets from other SCT platforms.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Verhoeckx K, Cotter P, López-Expósito I, Kleiveland C, Lea T, Mackie A, Requena T, Swiatecka D, Wichers H (2015) The impact of food bioactives on health: in vitro and ex vivo models. Springer, Cham
Shaikh RA, Zhong J, Lyu M, Lin S, Keskin D, Zhang G, Chitkushev L, Brusic V (2019) Classification of five cell types from PBMC samples using single cell transcriptomics and artificial neural networks. In: 2019 IEEE international conference on bioinformatics and biomedicine (BIBM). IEEE, pp 2207–2213
Baine MJ, Chakraborty S, Smith LM, Mallya K, Sasson AR, Brand RE, Batra SK (2011) Transcriptional profiling of peripheral blood mononuclear cells in pancreatic cancer patients identifies novel genes with potential diagnostic utility. PLoS One 6(2):e17014
Wang W-S, Liu L-X, Li G-P, Chen Y, Li C-Y, ** D-Y, Wang X-L (2013) Combined serum CA19-9 and miR-27a-3p in peripheral blood mononuclear cells to diagnose pancreatic cancer combined CA19-9 and miR-27a-3p to diagnose pancreatic cancer. Cancer Prev Res 6(4):331–338
Scott MK, Quinn K, Li Q, Carroll R, Warsinske H, Vallania F, Chen S, Carns MA, Aren K, Sun J (2019) Increased monocyte count as a cellular biomarker for poor outcomes in fibrotic diseases: a retrospective, multicentre cohort study. Lancet Respir Med 7(6):497–508
El-Awady MK, Ismail SM, El-Sagheer M, Sabour YA, Amr KS, Zaki EA (1999) Assay for hepatitis C virus in peripheral blood mononuclear cells enhances sensitivity of diagnosis and monitoring of HCV-associated hepatitis. Clin Chim Acta 283(1–2):1–14
Monaco G, Lee B, Xu W, Mustafah S, Hwang YY, Carre C, Burdin N, Visan L, Ceccarelli M, Poidinger M (2019) RNA-Seq signatures normalized by mRNA abundance allow absolute deconvolution of human immune cell types. Cell Rep 26(6):1627–40.e1627
Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7(3):562–578
Yang L, Zhang Y, Mitic N, Keskin DB, Zhang GL, Chitkushev L, Brusic V (2020) Single-cell mRNA Profiles in PBMC. In: 2020 IEEE international conference on bioinformatics and biomedicine (BIBM). IEEE, pp 1318–1323
Bakken T, Cowell L, Aevermann BD, Novotny M, Hodge R, Miller JA, Lee A, Chang I, McCorrison J, Pulendran B (2017) Cell type discovery and representation in the era of high-content single cell phenoty**. BMC Bioinf 18(17):7–16
Arendt D, Musser JM, Baker CV, Bergman A, Cepko C, Erwin DH, Pavlicev M, Schlosser G, Widder S, Laubichler MD (2016) The origin and evolution of cell types. Nat Rev Genet 17(12):744–757
Morris SA (2019) The evolving concept of cell identity in the single cell era. Development 146(12):dev169748
Kim HJ, Tam PP, Yang P (2021) Defining cell identity beyond the premise of differential gene expression. Cell Regen 10(1):1–3
Savulescu AF, Jacobs C, Negishi Y, Davignon L, Mhlanga MM (2020) Pinpointing cell identity in time and space. Front Mol Biosci 7:209
Lähnemann D, Köster J, Szczurek E, McCarthy DJ, Hicks SC, Robinson MD, Vallejos CA, Campbell KR, Beerenwinkel N, Mahfouz A (2020) Eleven grand challenges in single-cell data science. Genome Biol 21(1):1–35
Lin X, Zhong J, Lyu M, Lin S, Keskin DB, Zhang G, Brusic V, Chitkushev LT (2020) Artificial neural network system for cell classification using single cell RNA expression. In: 2020 IEEE international conference on bioinformatics and biomedicine (BIBM). IEEE, pp 1253–1257
Lyu M, Zhang Y, Yang L, Lin X, Li Y, ** H, Bellotti AG, Mitic N, Brusic V (2021) PBMC cell classification from single cell mRNA expression by artificial neural networks, profiles, gene markers, and protein markers. In: 2021 IEEE international conference on bioinformatics and biomedicine (BIBM). IEEE, pp 3285–3290
Deng Y, Bao F, Dai Q, Wu LF, Altschuler SJ (2019) Scalable analysis of cell-type composition from single-cell transcriptomics using deep recurrent learning. Nat Methods 16(4):311–314
Karaiskos N, Rahmatollahi M, Boltengagen A, Liu H, Hoehne M, Rinschen M, Schermer B, Benzing T, Rajewsky N, Kocks C (2018) A single-cell transcriptome atlas of the mouse glomerulus. J Am Soc Nephrol 29(8):2060–2068
Rendeiro AF, Krausgruber T, Fortelny N, Zhao F, Penz T, Farlik M, Schuster LC, Nemc A, Tasnády S, Réti M (2020) Chromatin map** and single-cell immune profiling define the temporal dynamics of ibrutinib response in CLL. Nat Commun 11(1):1–14
Ding J, Adiconis X, Simmons SK, Kowalczyk MS, Hession CC, Marjanovic ND, Hughes TK, Wadsworth MH, Burks T, Nguyen LT (2020) Systematic comparison of single-cell and single-nucleus RNA-sequencing methods. Nat Biotechnol 38(6):737–746
Zhang Y, Luning Y, Brusic V (2020) Automation of gene expression profile analysis in single cell data. In: 2020 IEEE international conference on bioinformatics and biomedicine (BIBM). IEEE, pp 1329–1334
Birney E, Andrews TD, Bevan P, Caccamo M, Chen Y, Clarke L, Coates G, Cuff J, Curwen V, Cutts T (2004) An overview of Ensembl. Genome Res 14(5):925–928
Lun AT, McCarthy DJ, Marioni JC (2016) A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Res 5:2122
Agarwal D, Sandor C, Volpato V, Caffrey TM, Monzón-Sandoval J, Bowden R, Alegre-Abarrategui J, Wade-Martins R, Webber C (2020) A single-cell atlas of the human substantia nigra reveals cell-specific pathways associated with neurological disorders. Nat Commun 11(1):1–11
Liu W, Venugopal S, Majid S, Ahn IS, Diamante G, Hong J, Yang X, Chandler SH (2020) Single-cell RNA-seq analysis of the brainstem of mutant SOD1 mice reveals perturbed cell types and pathways of amyotrophic lateral sclerosis. Neurobiol Dis 141:104877
Vicidomini R, Nguyen TH, Choudhury SD, Brody T, Serpe M (2021) Assembly and exploration of a single cell atlas of the drosophila larval ventral cord. Identification of rare cell types. Curr Protoc 1(2):e37
Luning Y, Zhang Y, Mitic N, Keskin DB, Zhang GL, Chitkushev L, Brusic V (2020) Prediction of PBMC cell types using scRNAseq reference profiles. In: 2020 IEEE international conference on bioinformatics and biomedicine (BIBM). IEEE, pp 1324–1328
Zhong J, Shaikh RA, Haoguo W, **n L, Zhiwei C, Chitkushev LT, Zhang G, Keskin DB, Brusic V (2020) Classification of PBMC cell types using scRNAseq, ANN, and incremental learning. In: 2020 IEEE international conference on bioinformatics and biomedicine (BIBM). IEEE, pp 1351–1355
Acknowledgments
DK received funding from the Division of Cancer Epidemiology and Genetics, National Cancer Institute (R21 CA216772-01A1) and from National Cancer Institute (SPORE-2P50CA101942-11A1). VB received Ningbo Municipal Bureau of Science and Technology Grant (2019F1028).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Lyu, M. et al. (2023). Protocol for Classification Single-Cell PBMC Types from Pathological Samples Using Supervised Machine Learning. In: Reche, P.A. (eds) Computational Vaccine Design. Methods in Molecular Biology, vol 2673. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-3239-0_4
Download citation
DOI: https://doi.org/10.1007/978-1-0716-3239-0_4
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-3238-3
Online ISBN: 978-1-0716-3239-0
eBook Packages: Springer Protocols