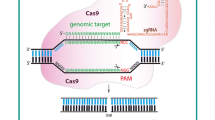
Abstract
Previous studies of gene function rely on the existing natural genetic variation or on induction of mutations by physical or chemical mutagenesis. The availability of alleles in nature, and random mutagenesis induced by physical or chemical means, limits the depth of research. The CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9) system provides the means to rapidly modify genomes in a precise and predictable way, making it possible to modulate gene expression and modify the epigenome. Barley is the most appropriate model species for functional genomic analysis of common wheat. Therefore, the genome editing system of barley is very important for the study of wheat gene function. Here we detail a protocol for barley gene editing. The effectiveness of this method has been confirmed in our previous published studies.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Bak RO, Gomez-Ospina N, Porteus MH (2018) Gene editing on center stage. Trends Genet 34(8):600–611
Woolf TM (1998) Therapeutic repair of mutated nucleic acid sequences. Nat Biotechnol 16(4):341–344
Voytas DF, Gao C (2014) Precision genome engineering and agriculture: opportunities and regulatory challenges. PLoS Biol 12(6):e1001877
**ng HL, Dong L, Wang ZP et al (2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol 14(1):327
Santamaria ME, Diaz-Mendoza M, Perez-Herguedas D et al (2018) Overexpression of HvIcy6 in barley enhances resistance against Tetranychus urticae and entails partial transcriptomic reprogramming. Int J Mol Sci 19(3):697
Ferdous J, Whitford R, Nguyen M et al (2017) Drought-inducible expression of Hv-miR827 enhances drought tolerance in transgenic barley. Funct Integr Genomics 17(2–3):279–292
Chen J, Liu C, Shi B et al (2017) Overexpression of HvHGGT enhances tocotrienol levels and antioxidant activity in Barley. J Agric Food Chem 65(25):5181–5187
Harwood WA (2014) A protocol for high-throughput Agrobacterium-mediated barley transformation. Methods Mol Biol 1099:251–260
Yang Q, Li S, Li X et al (2019) Expression of the high molecular weight glutenin 1Ay gene from Triticum urartu in barley. Transgenic Res 28(2):225–235
Hinchliffe A, Harwood WA (2019) Agrobacterium-mediated transformation of barley immature embryos. Methods Mol Biol 1900:115–126
Magoč T, Salzberg SL (2011) FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27(21):2957–2963
Jia H, Zhang Y, Orbović V et al (2017) Genome editing of the disease susceptibility gene Cs LOB 1 in citrus confers resistance to citrus canker. Plant Biotechnol J 15(7):817–823
Semenova E, Jore MM, Datsenko KA et al (2011) Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc Natl Acad Sci 108(25):10098–10103
Wiedenheft B, van Duijn E, Bultema JB et al (2011) RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc Natl Acad Sci 108(25):10092–10097
Gritz L, Davies J (1983) Plasmid-encoded hygromycin B resistance: the sequence of hygromycin B phosphotransferase gene and its expression in Escherichia coli and Saccharomyces cerevisiae. Gene 25(2–3):179–188
Wang MB, Waterhouse PM (1997) A rapid and simple method of assaying plants transformed with hygromycin or PPT resistance genes. Plant Mol Biol Rep 15(3):209–215
Naito Y, Hino K, Bono H et al (2015) CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics 31:1120–1123
Yang Q, Ding JJ, Feng XQ et al (2022) Editing of the starch synthase IIa gene led to transcriptomic andmetabolomic changes and high amylose starch in barley. Carbohydr Polym 285:119238
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Jiang, Q., Yang, Q., Harwood, W., Tang, H., Wei, Y., Zheng, Y. (2023). A CRISPR/Cas9 Protocol for Target Gene Editing in Barley. In: Yang, B., Harwood, W., Que, Q. (eds) Plant Genome Engineering. Methods in Molecular Biology, vol 2653. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-3131-7_18
Download citation
DOI: https://doi.org/10.1007/978-1-0716-3131-7_18
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-3130-0
Online ISBN: 978-1-0716-3131-7
eBook Packages: Springer Protocols