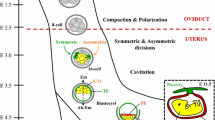
Abstract
Early cell specification in mammalian preimplantation embryos is an intricate cellular process that leads to coordinated spatial and temporal expression of specific genes. Proper segregation into the first two cell lineages, the inner cell mass (ICM) and the trophectoderm (TE), is imperative for develo** the embryo proper and the placenta, respectively. Somatic cell nuclear transfer (SCNT) allows the formation of a blastocyst containing both ICM and TE from a differentiated cell nucleus, which means that this differentiated genome must be reprogrammed to a totipotent state. Although blastocysts can be generated efficiently through SCNT, the full-term development of SCNT embryos is impaired mostly due to placental defects. In this review, we examine the early cell fate decisions in fertilized embryos and compare them to observations in SCNT-derived embryos, in order to understand if these processes are affected by SCNT and could be responsible for the low success of reproductive cloning.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Ma H, Morey R, O’Neil RC, He Y, Daughtry B, Schultz MD et al (2014) Abnormalities in human pluripotent cells due to reprogramming mechanisms. Nature 511:177–183
Yang X, Smith SL, Tian XC, Lewin H, Renard J-P, Wakayama T (2007) Nuclear reprogramming of cloned embryos and its implications for therapeutic cloning. Nat Genet 39:295–302
Heyman Y, Chavatte-Palmer P, LeBourhis D, Camous S, Vignon X, Renard JP (2002) Frequency and occurrence of late-gestation losses from cattle cloned embryos. Biol Reprod 66:6–13
Palmieri C, Loi P, Ptak G, Della Salda L (2008) Review Paper: A review of the pathology of abnormal placentae of somatic cell nuclear transfer clone pregnancies in cattle, sheep, and mice. Vet Pathol 45:865–880
Matoba S, Wang H, Jiang L, Lu F, Iwabuchi KA, Wu X et al (2018) Loss of H3K27me3 imprinting in somatic cell nuclear transfer embryos disrupts post-implantation development. Cell Stem Cell 23:343–354.e5
Cockburn K, Rossant J (2010) Making the blastocyst: lessons from the mouse. J Clin Invest 120:995–1003
Hillman N, Sherman MI, Graham C (1972) The effect of spatial arrangement on cell determination during mouse development. J Embryol Exp Morphol 28:263–278
Garner W, McLaren A (1974) Cell distribution in chimaeric mouse embryos before implantation. J Embryol Exp Morphol 32:495–503
Tabansky I, Lenarcic A, Draft RW, Loulier K, Keskin DB, Rosains J et al (2013) Developmental bias in cleavage-stage mouse blastomeres. Curr Biol 23:21–31
Casser E, Israel S, Witten A, Schulte K, Schlatt S, Nordhoff V et al (2017) Totipotency segregates between the sister blastomeres of two-cell stage mouse embryos. Sci Rep 7:1–15
Johnson WH, Loskutoff NM, Plante Y, Betteridge KJ (1995) Production of four identical calves by the separation of blastomeres from an in vitro derived four-cell embryo. Vet Rec 137:15–16
Willadsen SM (1980) The viability of early cleavage stages containing half the normal number of blastomeres in the sheep. J Reprod Fertil 59:357–362
Boiani M, Casser E, Fuellen G, Christians ES (2019) Totipotency continuity from zygote to early blastomeres: a model under revision. Reproduction 158:R49–R65
Stephenson RO, Rossant J, Tam PPL (2012) Intercellular interactions, position, and polarity in establishing blastocyst cell lineages and embryonic axes. Cold Spring Harb Perspect Biol 4:1–15
Sasaki H (2010) Mechanisms of trophectoderm fate specification in preimplantation mouse development. Develop Growth Differ 52:263–273
Plusa B, Frankenberg S, Chalmers A, Hadjantonakis A-K, Moore CA, Papalopulu N et al (2005) Downregulation of Par3 and aPKC function directs cells towards the ICM in the preimplantation mouse embryo. J Cell Sci 118:505–515
Anani S, Bhat S, Honma-Yamanaka N, Krawchuk D, Yamanaka Y (2014) Initiation of Hippo signaling is linked to polarity rather than to cell position in the pre-implantation mouse embryo. Development 141:2813–2824
Maître J-L, Turlier H, Illukkumbura R, Eismann B, Niwayama R, Nédélec F et al (2016) Asymmetric division of contractile domains couples cell positioning and fate specification. Nature 536:344–348
Korotkevich E, Niwayama R, Courtois A, Friese S, Berger N, Buchholz F et al (2017) The apical domain is required and sufficient for the first lineage segregation in the mouse embryo. Dev Cell 40:235–247.e7
Hirate Y, Hirahara S, Inoue KI, Suzuki A, Alarcon VB, Akimoto K et al (2013) Polarity-dependent distribution of angiomotin localizes hippo signaling in preimplantation embryos. Curr Biol 23:1181–1194
Yagi R, Kohn MJ, Karavanova I, Kaneko KJ, Vullhorst D, DePamphilis ML et al (2007) Transcription factor TEAD4 specifies the trophectoderm lineage at the beginning of mammalian development. Development 134:3827–3836
Nishioka N, Yamamoto S, Kiyonari H, Sato H, Sawada A, Ota M et al (2008) Tead4 is required for specification of trophectoderm in pre-implantation mouse embryos. Mech Dev 125:270–283
Nishioka N, Inoue KI, Adachi K, Kiyonari H, Ota M, Ralston A et al (2009) The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev Cell 16:398–410
Chi F, Sharpley MS, Nagaraj R, Roy SS, Banerjee U (2020) Glycolysis-independent glucose metabolism distinguishes TE from ICM fate during mammalian embryogenesis. Dev Cell 53:9–26.e4
Sakurai N, Takahashi K, Emura N, Hashizume T, Sawai K (2017) Effects of downregulating TEAD4 transcripts by RNA interference on early development of bovine embryos. J Reprod Dev 63:135–142
Akizawa H, Kobayashi K, Bai H, Takahashi M, Kagawa S, Nagatomo H et al (2018) Reciprocal regulation of TEAD4 and CCN2 for the trophectoderm development of the bovine blastocyst. Reproduction 155:563–571
Beck F, Erler T, Russell A, James R (1995) Expression of Cdx-2 in the mouse embryo and placenta: possible role in patterning of the extra-embryonic membranes. Dev Dyn 204:219–227
Strumpf D, Mao CA, Yamanaka Y, Ralston A, Chawengsaksophak K, Beck F et al (2005) Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development 132:2093–2102
Wu G, Gentile L, Fuchikami T, Sutter J, Psathaki K, Esteves TC et al (2010) Initiation of trophectoderm lineage specification in mouse embryos is independent of Cdx2. Development 137:4159–4169
Goissis MD, Cibelli JB (2014) Functional characterization of CDX2 during bovine preimplantation development in vitro. Mol Reprod Dev 81:962–970
Sakurai T, Bai H, Konno T, Ideta A, Aoyagi Y, Godkin JD et al (2010) Function of a transcription factor CDX2 beyond its trophectoderm lineage specification. Endocrinology 151:5873–5881
Bou G, Liu S, Sun M, Zhu J, Xue B, Guo J et al (2017) CDX2 is essential for cell proliferation and polarity in porcine blastocysts. Development 144:1296–1306
Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I et al (1998) Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95:379–391
Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R (2003) Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev 17:126–140
Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S et al (2003) Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 113:643–655
Loh Y-H, Wu Q, Chew J-L, Vega VB, Zhang W, Chen X et al (2006) The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet 38:431–440
Chew J, Loh Y, Zhang W, Chen X, Tam W, Yeap L et al (2006) Reciprocal transcriptional regulation of complex in embryonic stem cells reciprocal transcriptional regulation of Pou5f1 and Sox2 via the Oct4/Sox2 complex in embryonic stem cells. Mol Cell Biol 25:6031–6046
Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB et al (2008) Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133:1106–1117
Niwa H, Toyooka Y, Shimosato D, Strumpf D, Takahashi K, Yagi R et al (2005) Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell 123:917–929
Chen AE, Egli D, Niakan K, Deng J, Akutsu H, Yamaki M et al (2009) Optimal timing of inner cell mass isolation increases the efficiency of human embryonic stem cell derivation and allows generation of sibling cell lines. Cell Stem Cell 4:103–106
Cao S, Wang F, Chen Z, Liu Z, Mei C, Wu H et al (2009) Isolation and culture of primary bovine embryonic stem cell colonies by a novel method. J Exp Zool Part A Ecol Genet Physiol 311:368–376
Adjaye J, Huntriss J, Herwig R, BenKahla A, Brink TC, Wierling C et al (2005) Primary differentiation in the human blastocyst: comparative molecular portraits of inner cell mass and trophectoderm cells. Stem Cells 23:1514–1525
Kuijk EW, Du Puy L, Van Tol HT, Oei CHY, Haagsman HP, Colenbrander B et al (2008) Differences in early lineage segregation between mammals. Dev Dyn 237:918–927
Berg DK, Smith CS, Pearton DJ, Wells DN, Broadhurst R, Donnison M et al (2011) Trophectoderm lineage determination in cattle. Dev Cell 20:244–255
Guo G, Huss M, Tong GQ, Wang C, Li Sun L, Clarke ND et al (2010) Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev Cell 18:675–685
Wicklow E, Blij S, Frum T, Hirate Y, Lang R, Sasaki H et al (2014) HIPPO pathway members restrict SOX2 to the inner cell mass where it promotes ICM fates in the mouse blastocyst. PLoS Genet 10:e1004618
Frum T, Watts JL, Ralston A (2019) TEAD4, YAP1 and WWTR1 prevent the premature onset of pluripotency prior to the 16-cell stage. Development 146:dev179861. https://doi.org/10.1242/dev.179861
Goissis MD, Cibelli JB (2014) Functional characterization of SOX2 in bovine preimplantation embryos. Biol Reprod 90:30–30
Plachta N, Bollenbach T, Pease S, Fraser SE, Pantazis P (2011) Oct4 kinetics predict cell lineage patterning in the early mammalian embryo. Nat Cell Biol 13:117–123
Goolam M, Scialdone A, Graham SJL, Voet T, Marioni JC, Zernicka-goetz M et al (2016) Heterogeneity in Oct4 and Sox2 targets biases cell fate in 4-cell mouse embryos article heterogeneity in Oct4 and Sox2 targets biases cell fate in 4-cell mouse embryos. Cell 165:61–74
White MD, Angiolini JF, Alvarez YD, Kaur G, Zhao ZW, Mocskos E et al (2016) Long-lived binding of Sox2 to DNA predicts cell fate in the four-cell mouse embryo. Cell 165:75–87
Waksmundzka M, Wiśniewska A, Maleszewski M (2006) Allocation of cells in mouse blastocyst is not determined by the order of cleavage of the first two blastomeres. Biol Reprod 75:582–587
Kurotaki Y, Hatta K, Nakao K, Nabeshima YI, Fujimori T (2007) Blastocyst axis is specified independently of early cell lineage but aligns with the ZP shape. Science 316:719–723
Sepulveda-Rincon LP, Dube D, Adenot P, Laffont L, Ruffini S, Gall L et al (2016) Random allocation of blastomere descendants to the trophectoderm and ICM of the bovine blastocyst. Biol Reprod 95:123–123
Beyer TA, Weiss A, Khomchuk Y, Huang K, Ogunjimi AA, Varelas X et al (2013) Switch enhancers interpret TGF-β and hippo signaling to control cell fate in human embryonic stem cells. Cell Rep 5:1611–1624
Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K et al (2003) The homeoprotein nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113:631–642
Gardner RL, Rossant J (1979) Investigation of the fate of 4–5 day post-coitum mouse inner cell mass cells by blastocyst injection. J Embryol Exp Morphol 52:141–152
Gardner RL (1982) Investigation of cell lineage and differentiation in the extraembryonic endoderm of the mouse embryo. J Embryol Exp Morphol 68:175–198
Chazaud C, Yamanaka Y, Pawson T, Rossant J (2006) Early lineage segregation between epiblast and primitive endoderm in mouse blastocysts through the Grb2-MAPK pathway. Dev Cell 10:615–624
Plusa B, Piliszek A, Frankenberg S, Artus J, Hadjantonakis A-K (2008) Distinct sequential cell behaviours direct primitive endoderm formation in the mouse blastocyst. Development 135:3081–3091
Kang M, Piliszek A, Artus J, Hadjantonakis A-K (2013) FGF4 is required for lineage restriction and salt-and-pepper distribution of primitive endoderm factors but not their initial expression in the mouse. Development 140:267–279
Arman E, Haffner-Krausz R, Chen Y, Heath JK, Lonai P (1998) Targeted disruption of fibroblast growth factor (FGF) receptor 2 suggests a role for FGF signaling in pregastrulation mammalian development. Proc Natl Acad Sci U S A 95:5082–5087
Yamanaka Y, Lanner F, Rossant J (2010) FGF signal-dependent segregation of primitive endoderm and epiblast in the mouse blastocyst. Development 137:715–724
Frankenberg S, Gerbe F, Bessonnard S, Belville C, Pouchin P, Bardot O et al (2011) Primitive endoderm differentiates via a three-step mechanism involving Nanog and RTK signaling. Dev Cell 21:1005–1013
Schrode N, Saiz N, Di Talia S, Hadjantonakis AK (2014) GATA6 levels modulate primitive endoderm cell fate choice and timing in the mouse blastocyst. Dev Cell 29:454–467. https://doi.org/10.1016/j.devcel.2014.04.011
Bessonnard S, De Mot L, Gonze D, Barriol M, Dennis C, Goldbeter A et al (2014) Gata6, Nanog and Erk signaling control cell fate in the inner cell mass through a tristable regulatory network. Development 141:3637–3648
Kuijk EW, van Tol LTA, van de Velde H, Wubbolts R, Welling M, Geijsen N et al (2012) The roles of FGF and MAP kinase signaling in the segregation of the epiblast and hypoblast cell lineages in bovine and human embryos. Development 139:871–882
Canizo JR, Rivolta AEY, Echegaray CV, Suvá M, Alberio V, Aller JF et al (2019) A dose-dependent response to MEK inhibition determines hypoblast fate in bovine embryos. BMC Dev Biol 9:1–13
Ortega MS, Kelleher AM, O’Neil E, Benne J, Cecil R, Spencer TE (2020) NANOG is required to form the epiblast and maintain pluripotency in the bovine embryo. Mol Reprod Dev 87:152–160
Atlasi Y, Stunnenberg HG (2017) The interplay of epigenetic marks during stem cell differentiation and development. Nat Rev Genet 18:643–658
McLay DW, Clarke HJ (2003) Remodelling the paternal chromatin at fertilization in mammals. Reproduction 125:625–633
Santos F, Dean W (2004) Epigenetic reprogramming during early development in mammals. Reproduction 127:643–651
Saitou M, Kagiwada S, Kurimoto K (2012) Epigenetic reprogramming in mouse pre-implantation development and primordial germ cells. Development 139:15–31
Santos F, Hendrich B, Reik W, Dean W (2002) Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol 241:172–182
Nakanishi MO, Hayakawa K, Nakabayashi K, Hata K, Shiota K, Tanaka S (2012) Trophoblast-specific DNA methylation occurs after the segregation of the trophectoderm and inner cell mass in the mouse peri-implantation embryo. Epigenetics 7:173–182
Gaspar-Maia A, Alajem A, Meshorer E, Ramalho-Santos M (2011) Open chromatin in pluripotency and reprogramming. Nat Rev Mol Cell Biol 12:36–47
Beaujean N (2014) Histone post-translational modifications in preimplantation mouse embryos and their role in nuclear architecture. Mol Reprod Dev 81:100–112
Liu H, Kim JM, Aoki F (2004) Regulation of histone H3 lysine 9 methylation in oocytes and early pre-implantation embryos. Development 131:2269–2280
Lepikhov K, Walter J (2004) Differential dynamics of histone H3 methylation at positions K4 and K9 in the mouse zygote. BMC Dev Biol 4:2–6
Kourmouli N, Jeppesen P, Mahadevhaiah S, Burgoyne P, Wu R, Gilbert DM et al (2004) Heterochromatin and tri-methylated lysine 20 of histone H4 in animals. J Cell Sci 117:2491–2501
Daujat S, Weiss T, Mohn F, Lange UC, Ziegler-Birling C, Zeissler U et al (2009) H3K64 trimethylation marks heterochromatin and is dynamically remodeled during developmental reprogramming. Nat Struct Mol Biol 16:777–781
Wongtawan T, Taylor JE, Lawson KA, Wilmut I, Pennings S (2011) Histone H4K20me3 and HP1α are late heterochromatin markers in development, but present in undifferentiated embryonic stem cells. J Cell Sci 124:1878–1890
Ahmed K, Dehghani H, Rugg-Gunn P, Fussner E, Rossant J, Bazett-Jones DP (2010) Global chromatin architecture reflects pluripotency and lineage commitment in the early mouse embryo. PLoS One 5:0010531
Liu X, Wang C, Liu W, Li J, Li C, Kou X et al (2016) Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature 537:558–562
Erhardt S, Su IH, Schneider R, Barton S, Bannister AJ, Perez-Burgos L et al (2003) Consequences of the depletion of zygotic and embryonic enhancer of zeste 2 during preimplantation mouse development. Development 130:4235–4248
Dahl JA, Reiner AH, Klungland A, Wakayama T, Collas P (2010) Histone H3 lysine 27 methylation asymmetry on developmentally-regulated promoters distinguish the first two lineages in mouse preimplantation embryos. PLoS One 5:0009150
VerMilyea MD, O’Neill LP, Turner BM (2009) Transcription-independent heritability of induced histone modifications in the mouse preimplantation embryo. PLoS One 4:e6086
Torres-Padilla ME, Parfitt DE, Kouzarides T, Zernicka-Goetz M (2007) Histone arginine methylation regulates pluripotency in the early mouse embryo. Nature 445:214–218
Burton A, Muller J, Tu S, Padilla-Longoria P, Guccione E, Torres-Padilla ME (2013) Single-cell profiling of epigenetic modifiers identifies PRDM14 as an inducer of cell fate in the mammalian embryo. Cell Rep 5:687–701
Payer B, Lee JT (2008) X chromosome dosage compensation: how mammals keep the balance. Annu Rev Genet 42:733–772
Wutz A (2011) Gene silencing in X-chromosome inactivation: advances in understanding facultative heterochromatin formation. Nat Rev Genet 12:542–553
Mak W, Nesterova TB, De Napoles M, Appanah R, Yamanaka S, Otte AP et al (2004) Reactivation of the paternal X chromosome in early mouse embryos. Science 303:666–669
Silva J, Nichols J, Theunissen TW, Guo G, van Oosten AL, Barrandon O et al (2009) Nanog is the gateway to the pluripotent ground state. Cell 138:722–737
Petropoulos S, Edsgärd D, Reinius B, Deng Q, Panula SP, Codeluppi S et al (2016) Single-cell RNA-Seq reveals lineage and X chromosome dynamics in human preimplantation embryos. Cell 165:1012–1026
Vallot C, Patrat C, Collier AJ, Huret C, Casanova M, Liyakat Ali TM et al (2017) XACT noncoding RNA competes with XIST in the control of X chromosome activity during human early development. Cell Stem Cell 20:102–111
Okamoto I, Patrat C, Thépot D, Peynot N, Fauque P, Daniel N et al (2011) Eutherian mammals use diverse strategies to initiate X-chromosome inactivation during development. Nature 472:370–374
Ferreira AR, Machado GM, Diesel TO, Carvalho JO, Rumpf R, Melo EO et al (2010) Allele-specific expression of the MAOA gene and X chromosome inactivation in in vitro produced bovine embryos. Mol Reprod Dev 77:615–621
Yu B, van Tol HTA, Stout TAE, Roelen BAJ (2020) Initiation of X chromosome inactivation during bovine embryo development. Cell 9:1016
Briggs R, King TJ (1952) Transplantation of living nuclei from blastula cells into enucleated frogs’ eggs. Proc Natl Acad Sci 38:455–463
Gurdon JB, Melton DA (2008) Nuclear reprogramming in cells. Science 322:1811–1815
Wilmut I, Schnieke AE, McWhir J, Kind AJ, Campbell KHS (1997) Viable offspring derived from fetal and adult mammalian cells. Nature 385:810–813
Matoba S, Zhang Y (2018) Somatic cell nuclear transfer reprogramming: mechanisms and applications. Cell Stem Cell 23:471–485
Wakayama T, Tabar V, Rodriguez I, Perry ACF, Studer L, Mombaerts P (2001) Differentiation of embryonic stem cell lines generated from adult somatic cells by nuclear transfer. Science 292:740–743
Tachibana M, Amato P, Sparman M, Gutierrez NM, Tippner-Hedges R, Ma H et al (2013) Human embryonic stem cells derived by somatic cell nuclear transfer. Cell 153:1228–1238
Everts RE, Chavatte-Palmer P, Razzak A, Hue I, Green C, Oliveira R et al (2008) Aberrant gene expression patterns in placentomes are associated with phenotypically normal and abnormal cattle cloned by somatic cell nuclear transfer. Physiol Genomics 33:65–77
Chavatte-Palmer P, Camous S, Jammes H, Le Cleac’h N, Guillomot M, RSF L (2012) Review: Placental perturbations induce the developmental abnormalities often observed in bovine somatic cell nuclear transfer. Placenta 33:S99–S104
Ao Z, Liu D, Zhao C, Yue Z, Shi J, Zhou R et al (2017) Birth weight, umbilical and placental traits in relation to neonatal loss in cloned pigs. Placenta 57:94–101
Loi P, Clinton M, Vackova I, Fulka J, Feil R, Palmieri C et al (2006) Placental abnormalities associated with post-natal mortality in sheep somatic cell clones. Theriogenology 65:1110–1121
Tanaka S, Oda M, Toyoshima Y, Wakayama T, Tanaka M, Hattori N et al (2001) Placentomegaly in cloned mouse concepti caused by expansion of the spongiotrophoblast layer. Biol Reprod 65:1813–1821
Lin J, Shi L, Zhang M, Yang H, Qin Y, Zhang J et al (2011) Defects in trophoblast cell lineage account for the impaired in vivo development of cloned embryos generated by somatic nuclear transfer. Cell Stem Cell 8:371–375
Wang X, Qu J, Li J, He H, Liu Z, Huan Y (2020) Epigenetic reprogramming during somatic cell nuclear transfer: recent progress and future directions. Front Genet 11:1–13
Van Soom A, Ysebaert MT, De Kruif A (1997) Relationship between timing of development, morula morphology, and cell allocation to inner cell mass and trophectoderm in in vitro-produced bovine embryos. Mol Reprod Dev 47:47–56
Leese HJ, Donnay I, Thompson JG (1998) Human assisted conception: a cautionary tale. Lessons from domestic animals. Hum Reprod 13:184–202
Van Soom A, Boerjan M, Ysebaert MT, De Kruif A (1996) Cell allocation to the inner cell mass and the trophectoderm in bovine embryos cultured in two different media. Mol Reprod Dev 45:171–182
Spielmann H, Jacob-Mueller U, Beckord W (1980) Immunosurgical studies on inner cell mass development in rat and mouse blastocysts before and during implantation in vitro. J Embryol Exp Morphol 60:255–269
Iwasaki S, Yoshiba N, Ushijima H, Watanabe S, Nakahara T (1990) Morphology and proportion of inner cell mass of bovine blastocysts fertilized in vitro and in vivo. J Reprod Fertil 90:279–284
Papaioannou VE, Ebert KM (1988) The preimplantation pig embryo: cell number and allocation to trophectoderm and inner cell mass of the blastocyst in vivo and in vitro. Development 102:793–803
Koo D-B, Kang Y-K, Choi Y-H, Park JS, Kim H-N, Oh KB et al (2002) Aberrant allocations of inner cell mass and trophectoderm cells in bovine nuclear transfer blastocysts. Biol Reprod 67:487–492
Ushijima H, Akiyama K, Tajima T (2008) Transition of cell numbers in bovine preimplantation embryos: in vivo collected and in vitro produced embryos. J Reprod Dev 54:239–243
Boiani M, Eckardt S, Leu NA, Schöler HR, McLaughlin KJ (2003) Pluripotency deficit in clones overcome by clone-clone aggregation: epigenetic complementation? EMBO J 22:5304–5312
Misica-Turner PM, Oback FC, Eichenlaub M, Wells DN, Oback B (2007) Aggregating embryonic but not somatic nuclear transfer embryos increases cloning efficiency in cattle. Biol Reprod 76:268–278
Akagi S, Yamaguchi D, Matsukawa K, Mizutani E, Hosoe M, Adachi N et al (2011) Developmental ability of somatic cell nuclear transferred embryos aggregated at the 8-cell stage or 16- to 32-cell stage in cattle. J Reprod Dev 57:500–506
Bortvin A, Eggan K, Skaletsky H, Akutsu H, Berry DL, Yanagimachi R et al (2003) Incomplete reactivation of Oct4-related genes in mouse embryos cloned from somatic nuclei. Development 130:1673–1680
Takahashi K, Yamanaka S (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126:663–676
Buganim Y, Faddah DA, Jaenisch R (2013) Mechanisms and models of somatic cell reprogramming. Nat Rev Genet 14:427–439
Wen D, Banaszynski LA, Liu Y, Geng F, Noh KM, **ang J et al (2014) Histone variant H3.3 is an essential maternal factor for oocyte reprogramming. Proc Natl Acad Sci U S A 111:7325–7330
Rodríguez-Alvarez L, Manriquez J, Velasquez A, Castro FO (2013) Constitutive expression of the embryonic stem cell marker OCT4 in bovine somatic donor cells influences blastocysts rate and quality after nucleus transfer. Vitr Cell Dev Biol Anim 49:657–667
Beyhan Z, Ross PJ, Iager AE, Kocabas AM, Cunniff K, Rosa GJ et al (2007) Transcriptional reprogramming of somatic cell nuclei during preimplantation development of cloned bovine embryos. Dev Biol 305:637–649
Pfister-Genskow M, Myers C, Childs LA, Lacson JC, Patterson T, Betthauser JM et al (2005) Identification of differentially expressed genes in individual bovine preimplantation embryos produced by nuclear transfer: improper reprogramming of genes required for development. Biol Reprod 72:546–555
Somers J, Smith C, Donnison M, Wells DN, Henderson H, McLeay L et al (2006) Gene expression profiling of individual bovine nuclear transfer blastocysts. Reproduction 131:1073–1084
Min B, Cho S, Park JS, Lee YG, Kim N, Kang YK (2015) Transcriptomic features of bovine blastocysts derived by somatic cell nuclear transfer. G3 Genes, Genomes, Genet 5:2527–2538
Sood TJ, Lagah SV, Mukesh M, Singla SK, Chauhan MS, Manik RS et al (2019) RNA sequencing and transcriptome analysis of buffalo (Bubalus bubalis) blastocysts produced by somatic cell nuclear transfer and in vitro fertilization. Mol Reprod Dev 86:1149–1167
Wuensch A, Habermann FA, Kurosaka S, Klose R, Zakhartchenko V, Reichenbach H-D et al (2007) Quantitative monitoring of pluripotency gene activation after somatic cloning in cattle. Biol Reprod 76:983–991
Arnold DR, Bordignon V, Lefebvre R, Murphy BD, Smith LC (2006) Somatic cell nuclear transfer alters peri-implantation trophoblast differentiation in bovine embryos. Reproduction 132:279–290
Rayon T, Menchero S, Rollán I, Ors I, Helness A, Crespo M et al (2016) Distinct mechanisms regulate Cdx2 expression in the blastocyst and in trophoblast stem cells. Sci Rep 6:1–10
Rayon T, Menchero S, Nieto A, Xenopoulos P, Crespo M, Cockburn K et al (2014) Notch and Hippo converge on Cdx2 to specify the trophectoderm lineage in the mouse blastocyst. Dev Cell 30:410–422
Fujii T, Moriyasu S, Hirayama H, Hashizume T, Sawai K (2010) Aberrant expression patterns of genes involved in segregation of inner cell mass and trophectoderm lineages in bovine embryos derived from somatic cell nuclear transfer. Cell Reprogram 12:617–625
Wu X, Song M, Yang X, Liu X, Liu K, Jiao C et al (2016) Establishment of bovine embryonic stem cells after knockdown of CDX2. Sci Rep 6:1–12
Munsie MJ, Michalska AE, O’Brien CM, Trounson AO, Pera MF, Mountford PS (2000) Isolation of pluripotent embryonic stem cells from reprogrammed adult mouse somatic cell nuclei. Curr Biol 10:989–992
Wakayama S, Ohta H, Kishigami S, Van Thuan N, Hikichi T, Mizutani E et al (2005) Establishment of male and female nuclear transfer embryonic stem cell lines from different mouse strains and tissues. Biol Reprod 72:932–936
Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VSS et al (1998) Embryonic stem cell lines derived from human blastocysts. Science 282:1145–1147
Rielland M, Brochard V, Lacroix MC, Renard JP, Jouneau A (2009) Early alteration of the self-renewal/differentiation threshold in trophoblast stem cells derived from mouse embryos after nuclear transfer. Dev Biol 334:325–334
Oda M, Tanaka S, Yamazaki Y, Ohta H, Iwatani M, Suzuki M et al (2009) Establishment of trophoblast stem cell lines from somatic cell nuclear-transferred embryos. Proc Natl Acad Sci U S A 106:16293–16297
Xue F, Tian XC, Du F, Kubota C, Taneja M, Dinnyes A et al (2002) Aberrant patterns of X chromosome inactivation in bovine clones. Nat Genet 31:216–220
Williams LH, Kalantry S, Starmer J, Magnuson T (2011) Transcription precedes loss of **st coating and depletion of H3K27me3 during X-chromosome reprogramming in the mouse inner cell mass. Development 138:2049–2057
Muñoz M, Rodríguez A, De Frutos C, Caamaño JN, Díez C, Facal N et al (2008) Conventional pluripotency markers are unspecific for bovine embryonic-derived cell-lines. Theriogenology 69:1159–1164
Simmet K, Zakhartchenko V, Philippou-Massier J, Blum H, Klymiuk N, Wolf E (2018) OCT4/POU5F1 is required for NANOG expression in bovine blastocysts. Proc Natl Acad Sci 115:2770–2775
Inoue K, Kohda T, Sugimoto M, Sado T, Ogonuki N, Matoba S et al (2010) Impeding **st expression from the active X chromosome improves mouse somatic cell nuclear transfer. Science 330:496–499
Ruan D, Peng J, Wang X, Ouyang Z, Zou Q, Yang Y et al (2018) XIST derepression in active X chromosome hinders pig somatic cell nuclear transfer. Stem Cell Rep 10:494–508
Matoba S, Inoue K, Kohda T, Sugimoto M, Mizutani E, Ogonuki N et al (2011) RNAi-mediated knockdown of **st can rescue the impaired postimplantation development of cloned mouse embryos. Proc Natl Acad Sci 108:20621–20626
Alberto ML, Meirelles FV, Perecin F, Ambrósio CE, Favaron PO, Franciolli ALR et al (2012) Development of bovine embryos derived from reproductive techniques. Reprod Fertil Dev 25:907
Posfai E, Tam OH, Rossant J (2014) Mechanisms of pluripotency in vivo and in vitro, 1st edn. Elsevier Inc
Morgani S, Nichols J, Hadjantonakis AK (2017) The many faces of pluripotency: in vitro adaptations of a continuum of in vivo states. BMC Dev Biol 17:10–12
Daigneault BW, Rajput S, Smith GW, Ross PJ (2018) Embryonic POU5F1 is required for expanded bovine blastocyst formation. Sci Rep 8:7753
Fogarty NME, McCarthy A, Snijders KE, Powell BE, Kubikova N, Blakeley P et al (2017) Genome editing reveals a role for OCT4 in human embryogenesis. Nature 550:67–73
Tecirlioglu RT, Cooney MA, Lewis IM, Korfiatis NA, Hodgson R, Ruddock NT et al (2005) Comparison of two approaches to nuclear transfer in the bovine: hand-made cloning with modifications and the conventional nuclear transfer technique. Reprod Fertil Dev 17:573–585
Li Y, Li S, Dai Y, Du W, Zhao C, Wang L et al (2007) Nuclear reprogramming in embryos generated by the transfer of yak (Bos grunniens) nuclei into bovine oocytes and comparison with bovine-bovine SCNT and bovine IVF embryos. Theriogenology 67:1331–1338
Ross PJ, Rodriguez RM, Iager AE, Beyhan Z, Wang K, Ragina NP et al (2009) Activation of bovine somatic cell nuclear transfer embryos by PLCZ cRNA injection. Reproduction 137:427–437
Song BS, Kim JS, Kim CH, Han YM, Lee DS, Lee KK et al (2009) Prostacyclin stimulates embryonic development via regulation of the cAMP response element-binding protein-cyclo-oxygenase-2 signalling pathway in cattle. Reprod Fertil Dev 21:400–407
Cui X-S, Xu Y-N, Shen X-H, Zhang L-Q, Zhang J-B, Kim N-H (2011) Trichostatin A modulates apoptotic-related gene expression and improves embryo viability in cloned bovine embryos. Cell Reprogram 13:179–189
Hua S, Zhang H, Su JM, Zhang T, Quan FS, Liu J et al (2011) Effects of the removal of cytoplasm on the development of early cloned bovine embryos. Anim Reprod Sci 126:37–44
Su J, Wang Y, Li Y, Li R, Li Q, Wu Y et al (2011) Oxamflatin significantly improves nuclear reprogramming, blastocyst quality, and in vitro development of bovine SCNT embryos. PLoS One 6:0023805
Goissis MD, Suhr ST, Cibelli JB (2013) Effects of donor fibroblasts expressing OCT4 on bovine embryos generated by somatic cell nuclear transfer. Cell Reprogram 15:24–34
Chen H, Zhang L, Guo Z, Wang Y, He R, Qin Y et al (2015) Improving the development of early bovine somatic-cell nuclear transfer embryos by treating adult donor cells with vitamin C. Mol Reprod Dev 82:867–879
An Q, Peng W, Cheng Y, Lu Z, Zhou C, Zhang Y et al (2019) Melatonin supplementation during in vitro maturation of oocyte enhances subsequent development of bovine cloned embryos. J Cell Physiol 234:17370–17381
Chang HY, **e RX, Zhang L, Fu LZ, Zhang CT, Chen HH et al (2019) Overexpression of miR-101-2 in donor cells improves the early development of Holstein cow somatic cell nuclear transfer embryos. J Dairy Sci 102:4662–4673
Sah S, Sharma AK, Singla SK, Singh MK, Chauhan MS, Manik RS et al (2020) Effects of treatment with a microRNA mimic or inhibitor on the developmental competence, quality, epigenetic status and gene expression of buffalo (Bubalus bubalis) somatic cell nuclear transfer embryos. Reprod Fertil Dev 32:508–521
Hai T, Hao J, Wang L, Jouneau A, Zhou Q (2011) Pluripotency maintenance in mouse somatic cell nuclear transfer embryos and its improvement by treatment with the histone deacetylase inhibitor TSA. Cell Reprogram 13:47–56
Mizutani E, Torikai K, Wakayama S, Nagatomo H, Ohinata Y, Kishigami S et al (2016) Generation of cloned mice and nuclear transfer embryonic stem cell lines from urine-derived cells. Sci Rep 6:1–8
Kim G, Roy PK, Fang X, Hassan BMS, Cho J (2019) Improved preimplantation development of porcine somatic cell nuclear transfer embryos by caffeine treatment. J Vet Sci 20:1–12
Zhou Q, Yang SH, Ding CH, He XC, **e YH, Hildebrandt TB et al (2006) A comparative approach to somatic cell nuclear transfer in the rhesus monkey. Hum Reprod 21:2564–2571
Iager AE, Ragina NP, Ross PJ, Beyhan Z, Cunniff K, Rodriguez RM et al (2008) Trichostatin A improves histone acetylation in bovine somatic cell nuclear transfer early embryos. Cloning Stem Cells 10:371–379
Zhang J, Hao L, Wei Q, Zhang S, Cheng H, Zhai Y et al (2020) TET3 overexpression facilitates DNA reprogramming and early development of bovine SCNT embryos. Reproduction 160:379–391
Cervera RP, Martí-Gutiérrez N, Escorihuela E, Moreno R, Stojkovic M (2009) Trichostatin A affects histone acetylation and gene expression in porcine somatic cell nucleus transfer embryos. Theriogenology 72:1097–1110
Acknowledgments
The Sao Paulo State Research Foundation (FAPESP 2017/09576-3) and the National Council of Science and Technology (CNPq 408634/2018-9) currently fund Marcelo D. Goissis.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Goissis, M.D., Cibelli, J.B. (2023). Early Cell Specification in Mammalian Fertilized and Somatic Cell Nuclear Transfer Embryos. In: Moura, M.T. (eds) Somatic Cell Nuclear Transfer Technology . Methods in Molecular Biology, vol 2647. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-3064-8_3
Download citation
DOI: https://doi.org/10.1007/978-1-0716-3064-8_3
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-3063-1
Online ISBN: 978-1-0716-3064-8
eBook Packages: Springer Protocols