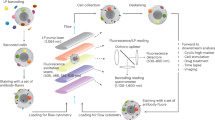
Abstract
Fluorescent cell barcoding (FCB) is a useful flow cytometric technique for high-throughput multiplexed analyses and can minimize technical variations after preliminary optimization and validation of protocols. To date, FCB is widely used for measurement of phosphorylation status of certain proteins, while it can be also employed for cellular viability assessment. In this chapter, we describe the protocol to perform FCB combined with viability assessment on lymphocytes and monocytes using manual and computational analysis. We also provide recommendations for FCB protocol optimization and validation for clinical sample analysis.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Krutzik PO, Nolan GP (2006) Fluorescent cell barcoding in flow cytometry allows high-throughput drug screening and signaling profiling. Nat Methods 3:361–368
Krutzik PO, Nolan GP (2003) Intracellular phospho-protein staining techniques for flow cytometry: monitoring single cell signaling events. Cytometry A 55:61–70
Krutzik PO, Crane JM, Clutter MR, Nolan GP (2008) High-content single-cell drug screening with phosphospecific flow cytometry. Nat Chem Biol 4:132–142
Kalland ME, Oberprieler NG, Vang T, Taskén K, Torgersen KM (2011) T cell-signaling network analysis reveals distinct differences between CD28 and CD2 costimulation responses in various subsets and in the MAPK pathway between resting and activated regulatory T cells. J Immunol 187:5233–5245
Spurgeon BE, Aburima A, Oberprieler NG, Taskén K, Naseem KM (2014) Multiplexed phosphospecific flow cytometry enables large-scale signaling profiling and drug screening in blood platelets. J Thromb Haemost 12:1733–1743
Davies R, Vogelsang P, Jonsson R, Appel S (2016) An optimized multiplex flow cytometry protocol for the analysis of intracellular signaling in peripheral blood mononuclear cells. J Immunol Methods 436:58–63
Tsai WL, Vian L, Giudice V, Kieltyka J, Liu C, Fonseca V, Gazaniga N, Gao S, Kajigaya S, Young NS, Biancotto A, Gadina M (2020) High throughput pSTAT signaling profiling by fluorescent cell barcoding and computational analysis. J Immunol Methods 477:112667
Manohar S, Shah P, Biswas S, Mukadam A, Joshi M, Viswanathan G (2019) Combining fluorescent cell barcoding and flow cytometry-based phospho-ERK1/2 detection at short time scales in adherent cells. Cytometry A 95(2):192–200
Stam J, Abdulahad W, Huitema MG et al (2011) Fluorescent cell barcoding as a tool to assess the age-related development of intracellular cytokine production in small amounts of blood from infants. PLoS One 6(10):e25690
Prussin C, Metcalfe DD (1995) Detection of intracytoplasmic cytokine using flow cytometry and directly conjugated anti-cytokine antibodies. J Immunol Methods 188:117–128
Giudice V, Feng X, Kajigaya S, Young NS, Biancotto A (2017) Optimization and standardization of fluorescent cell barcoding for multiplexed flow cytometric phenoty**. Cytometry A 91(7):694–703
Giudice V, Fantoni G, Biancotto A (2019) Fluorescent cell barcoding for Immunophenoty**. Methods Mol Biol 2032:53–68
Gao S, Wu Z, Arnold B, Diamond C, Batchu S, Giudice V, Alemu L, Raffo DQ, Feng X, Kajigaya S, Barrett J, Ito S, Young NS (2022) Single-cell RNA sequencing coupled to TCR profiling of large granular lymphocyte leukemia T cells. Nat Commun 13(1):1982
Lu Y, Biancotto A, Cheung F, Remmers E, Shah N, McCoy JP, Tsang JS (2016) Systematic analysis of cell-to-cell expression variation of T lymphocytes in a human cohort identifies aging and genetic associations. Immunity 45(5):1162–1175
Finak G, Langweiler M, Jaimes M et al (2016) Standardizing flow cytometry immunophenoty** analysis from the human immunophenoty** consortium. Sci Rep 6:20686
Saeys Y, Gassen SV, Lambrecht BN (2016) Computational flow cytometry: hel** to make sense of high-dimensional immunology data. Nat Rev Immunol 16(7):449–462
Aghaeepour N, Finak G; FlowCAP Consortium et al (2013) Critical assessment of automated flow cytometry data analysis techniques. Nat Methods 10(3):228–238
Hu Z, Jujjavarapu C, Hughey JJ et al (2018) MetaCyto: a tool for automated meta-analysis of mass and flow cytometry data. Cell Rep 24(5):1377–1388
Reisman BJ, Barone SM, Bachmann BO, Irish JM (2021) DebarcodeR increases fluorescent cell barcoding capacity and accuracy. Cytometry A 99(9):946–953
Perfetto SP, Chattopadhyay PK, Lamoreaux L, Nguyen R, Ambrozak D, Koup RA, Roederer M (2010) Amine-reactive dyes for dead cell discrimination in fixed samples. Curr Protoc Cytom Chapter 9:Unit 9.34
Perfetto SP, Chattopadhyay PK, Lamoreaux L, Nguyen R, Ambrozak D, Koup RA, Roederer M (2006) Amine reactive dyes: an effective tool to discriminate live and dead cells in polychromatic flow cytometry. J Immunol Methods 313(1–2):199–208
Johnson S, Nguyen V, Coder D (2013) Assessment of cell viability. Curr Protoc Cytom Chapter 9: Unit9.2
Lekishvili T, Campbell JJ (2018) Rapid comparative immunophenoty** of human mesenchymal stromal cells by a modified fluorescent cell barcoding flow cytometric assay. Cytometry A 93(9):905–915
Krutzik PO, Clutter MR, Trejo A, Nolan GP (2011) Fluorescent cell barcoding for multiplex flow cytometry. Curr Protoc Cytom Chapter 6:Unit 6.31
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Giudice, V., Fonseca, V., Selleri, C., Gadina, M. (2023). Cell Viability Multiplexing: Quantification of Cellular Viability by Barcode Flow Cytometry and Computational Analysis. In: Friedrich, O., Gilbert, D.F. (eds) Cell Viability Assays. Methods in Molecular Biology, vol 2644. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-3052-5_7
Download citation
DOI: https://doi.org/10.1007/978-1-0716-3052-5_7
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-3051-8
Online ISBN: 978-1-0716-3052-5
eBook Packages: Springer Protocols