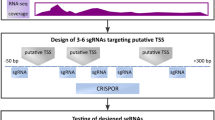
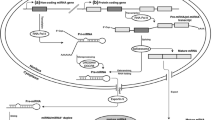
Abstract
Posttranscriptional silencing by microRNAs (miRNAs) is a critical constituent of eukaryotic gene regulation. miRNAs are short (~22 nt) noncoding RNAs capable of specifically targeting the miRNA-induced silencing complex (miRISC) to transcripts bearing a complementary miRNA response element (MRE). Although recent methodological advances have greatly improved our understanding of miRNA biogenesis and the mechanisms by which miRNAs repress their cognate targets, exploring the physiological relevance of direct miRNA-target interactions in vivo has remained an outstanding challenge. Here we describe the experimental protocol underlying a novel approach, which allows direct in situ interrogation of specific miRNA-MRE interactions by CRISPR/Cas9-mediated genome engineering (Bassett G et al., Nat Commun 5, 4640, 2014). In this instance, the CRISPR/Cas9 system is first used to catalyze homology-directed replacement of candidate MREs with molecular barcodes at endogenous loci. Subsequently, the effect of MRE mutation on transcript abundance (i.e., MRE activity) can be rapidly evaluated by routine quantitative PCR. This strategy enables functional investigation of a putative miRNA-target pair in a pool of transiently transfected cells, obviating the need for generation of clonal cell lines or transgenic animals. This protocol can be implemented in any cell line in less than 2 weeks and can readily be scaled up for multiplex studies. To facilitate the conceptual workflow underlying this strategy, we also describe a genome-wide resource for automated design and computational evaluation of CRISPR/Cas9 guide RNAs targeting all predicted MREs in various species (miR-CRISPR).
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Bassett AR, Azzam G, Wheatley L, Tibbit C, Rajakumar T, McGowan S, Stanger N, Ewels PA, Taylor S, Ponting CP, Liu JL, Sauka-Spengler T, Fulga TA (2014) Understanding functional miRNA-target interactions in vivo by site-specific genome engineering. Nat Commun 5:4640
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297
Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136:215–233
Lewis BP, Burge CB, Bartel DP (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120:15–20
Peterson SM, Thompson JA, Ufkin ML, Sathyanarayana P, Liaw L, Congdon CB (2014) Common features of microRNA target prediction tools. Front Genet 5:23
Li Y, Zhang Z (2015) Computational biology in microRNA. Wiley Interdiscip Rev RNA 6:435–452
Oulas A, Karathanasis N, Louloupi A, Pavlopoulos GA, Poirazi P, Kalantidis K, Iliopoulos I (2015) Prediction of miRNA targets. Methods Mol Biol 1269:207–229
Friedman RC, Farh KK, Burge CB, Bartel DP (2009) Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19:92–105
Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM (2005) Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433:769–773
Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N (2008) Widespread changes in protein synthesis induced by microRNAs. Nature 455:58–63
Xu G, Fewell C, Taylor C, Deng N, Hedges D, Wang X, Zhang K, Lacey M, Zhang H, Yin Q, Cameron J, Lin Z, Zhu D, Flemington EK (2010) Transcriptome and targetome analysis in MIR155 expressing cells using RNA-seq. RNA 16:1610–1622
Mansfield JH, Harfe BD, Nissen R, Obenauer J, Srineel J, Chaudhuri A, Farzan-Kashani R, Zuker M, Pasquinelli AE, Ruvkun G, Sharp PA, Tabin CJ, McManus MT (2004) MicroRNA-responsive ‘sensor’ transgenes uncover Hox-like and other developmentally regulated patterns of vertebrate microRNA expression. Nat Genet 36:1079–1083
Chi SW, Zang JB, Mele A, Darnell RB (2009) Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 460:479–486
Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M Jr, Jungkamp AC, Munschauer M, Ulrich A, Wardle GS, Dewell S, Zavolan M, Tuschl T (2010) Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141:129–141
Helwak A, Kudla G, Dudnakova T, Tollervey D (2013) Map** the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell 153:654–665
Agarwal V, Bell GW, Nam JW, Bartel DP (2015) Predicting effective microRNA target sites in mammalian mRNAs. elife 4:e05005
Sternberg SH, Doudna JA (2015) Expanding the Biologist’s toolkit with CRISPR-Cas9. Mol Cell 58:568–574
Maruyama T, Dougan SK, Truttmann MC, Bilate AM, Ingram JR, Ploegh HL (2015) Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol 33:538–542
Chu VT, Weber T, Wefers B, Wurst W, Sander S, Rajewsky K, Kuhn R (2015) Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat Biotechnol 33:543–548
Pyzocha NK, Ran FA, Hsu PD, Zhang F (2014) RNA-guided genome editing of mammalian cells. Methods Mol Biol 1114:269–277
Aricescu AR, Lu W, Jones EY (2006) A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr D Biol Crystallogr 62:1243–1250
Fraley SI, Hardick J, Jo Masek B, Athamanolap P, Rothman RE, Gaydos CA, Carroll KC, Wakefield T, Wang TH, Yang S (2013) Universal digital high-resolution melt: a novel approach to broad-based profiling of heterogeneous biological samples. Nucleic Acids Res 41:e175
Guschin DY, Waite AJ, Katibah GE, Miller JC, Holmes MC, Rebar EJ (2010) A rapid and general assay for monitoring endogenous gene modification. Methods Mol Biol 649:247–256. https://doi.org/10.1007/978-1-60761-753-2_15
Brinkman EK, Chen T, Amendola M, van Steensel B (2014) Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res 42(22):e168. https://doi.org/10.1093/nar/gku936
Wu Q, Ferry QRV, Baeumler TA, Michaels YS, Vitsios DM, Habib O, Arnold R, Jiang X, Maio S, Steinkraus BR, Tapia M, Piazza P, Xu N, Hollander GA, Milne TA, Kim JS, Enright AJ, Bassett AR, Fulga TA (2017) In situ functional dissection of RNA cis-regulatory elements by multiplex CRISPR-Cas9 genome engineering. Nat Commun 8(1):2109. https://doi.org/10.1038/s41467-017-00686-2
Acknowledgments
We thank Andrew Basset for critical comments on the manuscript and the Fulga lab for feedback and advice. Y.S.M. is supported by the Banting postdoctoral fellowship and the Michael Smith Foundation for Health Research Trainee award. Q.W. is supported by the Medical Research Council (WIMM Strategic Award, MRC #G0902418 to T.A.F.). T.A.F. was supported by the Medical Research Council (WIMM Strategic Award, MRC #G0902418), Biotechnology and Biological Sciences Research Council (Project Grants #BB/L010275/1 and #BB/N006550/1), and Welcome Trust ISSF Award (#105605/Z/14/Z).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Wu, Q., Michaels, Y.S., Fulga, T.A. (2023). Interrogation of Functional miRNA-Target Interactions by CRISPR/Cas9 Genome Engineering. In: Dalmay, T. (eds) MicroRNA Detection and Target Identification. Methods in Molecular Biology, vol 2630. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-2982-6_16
Download citation
DOI: https://doi.org/10.1007/978-1-0716-2982-6_16
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-2981-9
Online ISBN: 978-1-0716-2982-6
eBook Packages: Springer Protocols