Abstract
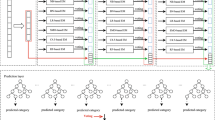
Pseudouridine is a ubiquitous RNA modification and plays a crucial role in many biological processes. However, it remains a challenging task to identify pseudouridine sites using expensive and time-consuming experimental research. To this end, we present Porpoise, a computational approach to identify pseudouridine sites from RNA sequence data. Porpoise builds on a stacking ensemble learning framework with several informative features and achieves competitive performance compared with state-of-the-art approaches. This protocol elaborates on step-by-step use and execution of the local stand-alone version and the webserver of Porpoise. In addition, we also provide a general machine learning framework that can help identify the optimal stacking ensemble learning model using different combinations of feature-based features. This general machine learning framework can facilitate users to build their pseudouridine predictors using their in-house datasets.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Basak A, Query CC (2014) A pseudouridine residue in the spliceosome core is part of the filamentous growth program in yeast. Cell Rep 8(4):966–973. https://doi.org/10.1016/j.celrep.2014.07.004
Carlile TM, Rojas-Duran MF, Zinshteyn B, Shin H, Bartoli KM, Gilbert WV (2014) Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 515(7525):143–146. https://doi.org/10.1038/nature13802
Charette M, Gray MW (2000) Pseudouridine in RNA: what, where, how, and why. IUBMB Life 49(5):341–351. https://doi.org/10.1080/152165400410182
Davis DR, Veltri CA, Nielsen L (1998) An RNA model system for investigation of pseudouridine stabilization of the codon-anticodon interaction in tRNALys, tRNAHis and tRNATyr. J Biomol Struct Dyn 15(6):1121–1132. https://doi.org/10.1080/07391102.1998.10509006
Jack K, Bellodi C, Landry DM, Niederer RO, Meskauskas A, Musalgaonkar S, Kopmar N, Krasnykh O, Dean AM, Thompson SR (2011) rRNA pseudouridylation defects affect ribosomal ligand binding and translational fidelity from yeast to human cells. Mol Cell 44(4):660–666. https://doi.org/10.1016/j.molcel.2011.09.017
Ma X, Zhao X, Yu YT (2003) Pseudouridylation (Ψ) of U2 snRNA in S. cerevisiae is catalyzed by an RNA-independent mechanism. EMBO J 22(8):1889–1897. https://doi.org/10.1038/sj.emboj.7600718
Mei Y, Liao J, Shen J, Yu L, Liu B, Liu L, Li R, Ji L, Dorsey S, Jiang Z (2012) Small nucleolar RNA 42 acts as an oncogene in lung tumorigenesis. Oncogene 31(22):2794–2804. https://doi.org/10.1038/onc.2011.449
Li X, Zhu P, Ma S, Song J, Bai J, Sun F, Yi C (2015) Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat Chem Biol 11(8):592–597. https://doi.org/10.1038/nchembio.1836
Li Y-H, Zhang G, Cui Q (2015) PPUS: a web server to predict PUS-specific pseudouridine sites. Bioinformatics 31(20):3362–3364. https://doi.org/10.1093/bioinformatics/btv366
Chen W, Tang H, Ye J, Lin H, Chou K-C (2016) iRNA-PseU: identifying RNA pseudouridine sites. Mol Ther Nucleic Acids 5:e332. https://doi.org/10.1038/mtna.2016.37
Bi Y, ** D, Jia C (2020) EnsemPseU: identifying pseudouridine sites with an ensemble approach. IEEE Access 8:79376–79382. https://doi.org/10.1109/ACCESS.2020.2989469
He J, Fang T, Zhang Z, Huang B, Zhu X, **ong Y (2018) PseUI: Pseudouridine sites identification based on RNA sequence information. BMC Bioinformatics 19(1):1–11. https://doi.org/10.1186/s12859-018-2321-0
Khan SM, He F, Wang D, Chen Y, Xu D (2020) MU-PseUDeep: a deep learning method for prediction of pseudouridine sites. Comput Struct Biotechnol J 18:1877–1883. https://doi.org/10.1016/j.csbj.2020.07.010
Liu K, Chen W, Lin H (2020) XG-PseU: an eXtreme gradient boosting based method for identifying pseudouridine sites. Mol Gen Genomics 295(1):13–21. https://doi.org/10.1007/s00438-019-01600-9
Lv Z, Zhang J, Ding H, Zou Q (2020) RF-PseU: a random forest predictor for RNA pseudouridine sites. Front Bioeng Biotechnol 8:134. https://doi.org/10.3389/fbioe.2020.00134
Tahir M, Tayara H, Chong KT (2019) iPseU-CNN: identifying RNA pseudouridine sites using convolutional neural networks. Mol Ther Nucleic Acids 16:463–470. https://doi.org/10.1016/j.omtn.2019.03.010
Song B, Tang Y, Wei Z, Liu G, Su J, Meng J, Chen K (2020) PIANO: a web server for pseudouridine-site (Ψ) identification and functional annotation. Front Genet 11:88. https://doi.org/10.3389/fgene.2020.00088
Song B, Chen K, Tang Y, Ma J, Meng J, Wei Z (2020) PSI-MOUSE: predicting mouse pseudouridine sites from sequence and genome-derived features. Evol Bioinform 16:1176934320925752. https://doi.org/10.1177/1176934320925752
Li F, Guo X, ** P, Chen J, **ang D, Song J, Coin LJM (2021) Porpoise: a new approach for accurate prediction of RNA pseudouridine sites. Brief Bioinform. https://doi.org/10.1093/bib/bbab245
Sun W-J, Li J-H, Liu S, Wu J, Zhou H, Qu L-H, Yang J-H (2016) RMBase: a resource for decoding the landscape of RNA modifications from high-throughput sequencing data. Nucleic Acids Res 44(D1):D259–D265. https://doi.org/10.1093/nar/gkv1036
Chen Z, Zhao P, Li F, Marquez-Lago TT, Leier A, Revote J, Zhu Y, Powell DR, Akutsu T, Webb GI, Chou KC, Smith AI, Daly RJ, Li J, Song J (2020) iLearn: an integrated platform and meta-learner for feature engineering, machine-learning analysis and modeling of DNA, RNA and protein sequence data. Brief Bioinform 21(3):1047–1057. https://doi.org/10.1093/bib/bbz041
Li F, Chen J, Leier A, Marquez-Lago T, Liu Q, Wang Y, Revote J, Smith AI, Akutsu T, Webb GI, Kurgan L, Song J (2020) DeepCleave: a deep learning predictor for caspase and matrix metalloprotease substrates and cleavage sites. Bioinformatics 36(4):1057–1065. https://doi.org/10.1093/bioinformatics/btz721
Li F, Leier A, Liu Q, Wang Y, **ang D, Akutsu T, Webb GI, Smith AI, Marquez-Lago T, Li J, Song J (2020) Procleave: predicting protease-specific substrate cleavage sites by combining sequence and structural information. Genomics Proteomics Bioinformatics 18(1):52–64. https://doi.org/10.1016/j.gpb.2019.08.002
Chen Z, Zhao P, Li C, Li F, **ang D, Chen YZ, Akutsu T, Daly RJ, Webb GI, Zhao Q, Kurgan L, Song J (2021) iLearnPlus: a comprehensive and automated machine-learning platform for nucleic acid and protein sequence analysis, prediction and visualization. Nucleic Acids Res 49(10):e60. https://doi.org/10.1093/nar/gkab122
Li F, Chen J, Ge Z, Wen Y, Yue Y, Hayashida M, Baggag A, Bensmail H, Song J (2021) Computational prediction and interpretation of both general and specific types of promoters in Escherichia coli by exploiting a stacked ensemble-learning framework. Brief Bioinform 22(2):2126–2140. https://doi.org/10.1093/bib/bbaa049
Liu Q, Chen J, Wang Y, Li S, Jia C, Song J, Li F (2021) DeepTorrent: a deep learning-based approach for predicting DNA N4-methylcytosine sites. Brief Bioinform 22(3):bbaa124. https://doi.org/10.1093/bib/bbaa124
Mei S, Li F, **ang D, Ayala R, Faridi P, Webb GI, Illing PT, Rossjohn J, Akutsu T, Croft NP, Purcell AW, Song J (2021) Anthem: a user customised tool for fast and accurate prediction of binding between peptides and HLA class I molecules. Brief Bioinform. https://doi.org/10.1093/bib/bbaa415
Zhu Y, Li F, **ang D, Akutsu T, Song J, Jia C (2020) Computational identification of eukaryotic promoters based on cascaded deep capsule neural networks. Brief Bioinform. https://doi.org/10.1093/bib/bbaa299
Chai D, Jia C, Zheng J, Zou Q, Li F (2021) Staem5: a novel computational approachfor accurate prediction of m5C site. Mol Ther Nucleic Acids 26:1027–1034. https://doi.org/10.1016/j.omtn.2021.10.012
Wang X, Li F, Xu J, Rong J, Webb GI, Ge Z, Li J, Song J (2022) ASPIRER: a new computational approach for identifying non-classical secreted proteins based on deep learning. Brief Bioinform. https://doi.org/10.1093/bib/bbac031
Li F, Guo X, **ang D, Pitt ME, Bainomugisa A, Coin LJ (2022) Computational analysis and prediction of PE_PGRS proteins using machine learning. Comput Struct Biotechnol J. https://doi.org/10.1016/j.csbj.2022.01.019
Li F, Dong S, Leier A, Han M, Guo X, Xu J, Wang X, Pan S, Jia C, Zhang Y (2022) Positive-unlabeled learning in bioinformatics and computational biology: a brief review. Brief Bioinform 23(1):bbab461. https://doi.org/10.1093/bib/bbab461
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Guo, X., Li, F., Song, J. (2023). Predicting Pseudouridine Sites with Porpoise. In: Oliveira, P.H. (eds) Computational Epigenomics and Epitranscriptomics. Methods in Molecular Biology, vol 2624. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-2962-8_10
Download citation
DOI: https://doi.org/10.1007/978-1-0716-2962-8_10
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-2961-1
Online ISBN: 978-1-0716-2962-8
eBook Packages: Springer Protocols