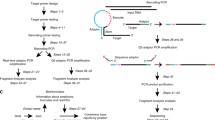
Abstract
The emergence of circulating DNA analysis in blood during the past decade has responded to the need for noninvasive alternatives to classical tissue biopsies. This has coincided with the development of techniques that allow the detection of low-frequency allele variants in clinical samples that typically carry very low amounts of fragmented DNA, such as plasma or FFPE samples. Enrichment of rare variants by nuclease-assisted mutant allele enrichment with overlap** probes (NaME-PrO) enables a more sensitive detection of mutations in tissue biopsy samples alongside standard qPCR detection assays. Such sensitivity is normally achieved by other more complex PCR methods, such as TaqMan qPCR and digital droplet PCR (ddPCR). Here we describe a workflow of mutation-specific nuclease-based enrichment combined with a SYBR Green real-time quantitative PCR detection method that provides comparable results to ddPCR. Using a PIK3CA mutation as an example, this combined workflow enables detection and accurate prediction of initial variant allele fraction in samples with a low mutant allele frequency (<1%) and could be applied flexibly to detect other mutations of interest.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Hagemann IS (2015) Chapter 1 – Overview of technical aspects and chemistries of next-generation sequencing. In: Kulkarni S, Pfeifer J (eds) Clinical genomics. Academic Press, Boston, pp 3–19. https://doi.org/10.1016/B978-0-12-404748-8.00001-0
Sabour L, Sabour M, Ghorbian S (2017) Clinical applications of next-generation sequencing in cancer diagnosis. Pathol Oncol Res 23:225. https://doi.org/10.1007/s12253-016-0124-z
Denis JA, Guillerm E, Coulet F, Larsen AK, Lacorte J-M (2017) The role of BEAMing and digital PCR for multiplexed analysis in molecular oncology in the era of next-generation sequencing. Mol Diagn Ther 21(6):587–600. https://doi.org/10.1007/s40291-017-0287-7
Krypuy M, Newnham GM, Thomas DM, Conron M, Dobrovic A (2006) High resolution melting analysis for the rapid and sensitive detection of mutations in clinical samples: KRAS codon 12 and 13 mutations in non-small cell lung cancer. BMC Cancer 6. https://doi.org/10.1186/1471-2407-6-295
Zhao J (2016) A sensitive and practical method to detect the T790M mutation in the epidermal growth factor receptor. Oncol Lett 11. https://doi.org/10.3892/ol.2016.4263
Olmedillas-López S, García-Arranz M, García-Olmo D (2017) Current and emerging applications of droplet digital PCR in oncology. Mol Diagn Ther 21(5):493–510. https://doi.org/10.1007/s40291-017-0278-8
Milbury CA, Li J, Liu P, Makrigiorgos GM (2011) COLD-PCR: improving the sensitivity of molecular diagnostics assays. Expert Rev Mol Diagn 11(2):159–169. https://doi.org/10.1586/erm.10.115
Ang D, O’Gara R, Schilling A, Beadling C, Warrick A, Troxell ML, Corless CL (2013) Novel method for PIK3CA mutation analysis: locked nucleic acid–PCR sequencing. J Mol Diagn 15(3):312–318. https://doi.org/10.1016/j.jmoldx.2012.12.005
Thierry AR (2014) Clinical validation of the detection of KRAS and BRAF mutations from circulating tumorDNA. Nat Med 20. https://doi.org/10.1038/nm.3511
Alvarez-Garcia V, Bartos C, Keraite I, Trivedi U, Brennan PM, Kersaudy-Kerhoas M, Gharbi K, Oikonomidou O, Leslie NR (2018) A simple and robust real-time qPCR method for the detection of PIK3CA mutations. Sci Rep 8(1):4290. https://doi.org/10.1038/s41598-018-22473-9
Gorgannezhad L, Umer M, Islam MN, Nguyen N-T, Shiddiky MJA (2018) Circulating tumor DNA and liquid biopsy: opportunities, challenges, and recent advances in detection technologies. Lab Chip 18(8):1174–1196. https://doi.org/10.1039/C8LC00100F
Ignatiadis M, Lee M, Jeffrey SS (2015) Circulating tumor cells and circulating tumor DNA: challenges and opportunities on the path to clinical utility. Clin Cancer Res 21(21):4786. https://doi.org/10.1158/1078-0432.CCR-14-1190
Wang J, Chang S, Li G, Sun Y (2017) Application of liquid biopsy in precision medicine: opportunities and challenges. Front Med 11(4):522–527. https://doi.org/10.1007/s11684-017-0526-7
Lennon NJ, Adalsteinsson VA, Gabriel SB (2016) Technological considerations for genome-guided diagnosis and management of cancer. Genome Med 8(1):112. https://doi.org/10.1186/s13073-016-0370-4
Song C, Liu Y, Fontana R, Makrigiorgos A, Mamon H, Kulke MH, Makrigiorgos GM (2016) Elimination of unaltered DNA in mixed clinical samples via nuclease-assisted minor-allele enrichment. Nucleic Acids Res 44(19):e146. https://doi.org/10.1093/nar/gkw650
Thierry AR, Mouliere F, El Messaoudi S, Mollevi C, Lopez-Crapez E, Rolet F, Gillet B, Gongora C, Dechelotte P, Robert B, Del Rio M, Lamy P-J, Bibeau F, Nouaille M, Loriot V, Jarrousse A-S, Molina F, Mathonnet M, Pezet D, Ychou M (2014) Clinical validation of the detection of KRAS and BRAF mutations from circulating tumor DNA. Nat Med 20:430. https://doi.org/10.1038/nm.3511
Keraite I, Alvarez-Garcia V, Garcia-Murillas I, Beaney M, Turner NC, Bartos C, Oikonomidou O, Kersaudy-Kerhoas M, Leslie NR (2020) PIK3CA mutation enrichment and quantitation from blood and tissue. Sci Rep 10(1):17082. https://doi.org/10.1038/s41598-020-74086-w
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25(4):402–408. https://doi.org/10.1006/meth.2001.1262
Cristiano S, Leal A, Phallen J, Fiksel J, Adleff V, Bruhm DC, Jensen S, Medina JE, Hruban C, White JR, Palsgrove DN, Niknafs N, Anagnostou V, Forde P, Naidoo J, Marrone K, Brahmer J, Woodward BD, Husain H, van Rooijen KL, Ørntoft MW, Madsen AH, van de Velde CJH, Verheij M, Cats A, Punt CJA, Vink GR, van Grieken NCT, Koopman M, Fijneman RJA, Johansen JS, Nielsen HJ, Meijer GA, Andersen CL, Scharpf RB, Velculescu VE (2019) Genome-wide cell-free DNA fragmentation in patients with cancer. Nature 570(7761):385–389. https://doi.org/10.1038/s41586-019-1272-6
Acknowledgments
The authors’ work develo** these methods was funded by the Chief Scientist Office (ETM/433) and by Medical Research Scotland (PhD-883-2015).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Keraite, I., Alvarez-Garcia, V., Leslie, N.R. (2023). Nuclease Enrichment and qPCR Detection of Rare Nucleotide Variants. In: Myers, M.B., Schandl, C.A. (eds) Clinical Applications of Nucleic Acid Amplification. Methods in Molecular Biology, vol 2621. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-2950-5_4
Download citation
DOI: https://doi.org/10.1007/978-1-0716-2950-5_4
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-2949-9
Online ISBN: 978-1-0716-2950-5
eBook Packages: Springer Protocols