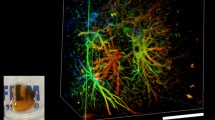
Abstract
While histological sections are the go-to method to analyze spatial information in tissues, three-dimensional information is fundamental to understand biological processes. Yet, the opaqueness of fixed samples prevents the visualization of large volumes of tissue. Tissue clearing allows to match the refractive indices of biological specimens with the mounting media, allowing light penetration and imaging of millimetric depths within tissues. We provide a detailed method to process whole brains for volumetric imaging in the African killifish Nothobranchius furzeri. We anticipate this method will facilitate the study of aging and regeneration of adult vertebrate organs.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Vieites-Prado A, Renier N (2021) Tissue clearing and 3D imaging in developmental biology. Development 148:dev199369
Brenna C, Simioni C, Varano G, Conti I, Costanzi E, Melloni M, Neri LM (2022) Optical tissue clearing associated with 3D imaging: application in preclinical and clinical studies. Histochem Cell Biol 157(5):497–511
Dodt HU, Leischner U, Schierloh A, Jahrling N, Mauch CP, Deininger K, Deussing JM, Eder M, Zieglgansberger W, Becker K (2007) Ultramicroscopy: three-dimensional visualization of neuronal networks in the whole mouse brain. Nat Methods 4:331–336
Almagro J, Messal HA, Zaw Thin M, van Rheenen J, Behrens A (2021) Tissue clearing to examine tumour complexity in three dimensions. Nat Rev Cancer 21:718–730
Chung K, Wallace J, Kim SY, Kalyanasundaram S, Andalman AS, Davidson TJ, Mirzabekov JJ, Zalocusky KA, Mattis J, Denisin AK et al (2013) Structural and molecular interrogation of intact biological systems. Nature 497:332–337
Kim SY, Cho JH, Murray E, Bakh N, Choi H, Ohn K, Ruelas L, Hubbert A, McCue M, Vassallo SL et al (2015) Stochastic electrotransport selectively enhances the transport of highly electromobile molecules. Proc Natl Acad Sci U S A 112:E6274–E6283
Park YG, Sohn CH, Chen R, McCue M, Yun DH, Drummond GT, Ku T, Evans NB, Oak HC, Trieu W et al (2018) Protection of tissue physicochemical properties using polyfunctional crosslinkers. Nat Biotechnol 37:73–83
Jordan T, Williams D, Criswell S, Wang Y (2019) Comparison of bleaching protocols utilizing hematoxylin and eosin stain and immunohistochemical proliferation marker MCM3 in pigmented melanomas. J Histotechnol 42:177–182
Shan QH, Qin XY, Zhou N, Huang C, Wang Y, Chen P, Zhou JN (2022) A method for ultrafast tissue clearing that preserves fluorescence for multimodal and longitudinal brain imaging. BMC Biol 20:77
Ueda HR, Erturk A, Chung K, Gradinaru V, Chedotal A, Tomancak P, Keller PJ (2020) Tissue clearing and its applications in neuroscience. Nat Rev Neurosci 21:61–79
Barykina NV, Karasev MM, Verkhusha VV, Shcherbakova DM (2022) Technologies for large-scale map** of functional neural circuits active during a user-defined time window. Prog Neurobiol 216:102290
Roy DS, Park YG, Kim ME, Zhang Y, Ogawa SK, DiNapoli N, Gu X, Cho JH, Choi H, Kamentsky L et al (2022) Brain-wide map** reveals that engrams for a single memory are distributed across multiple brain regions. Nat Commun 13:1799
Pang Z, Schafroth MA, Ogasawara D, Wang Y, Nudell V, Lal NK, Yang D, Wang K, Herbst DM, Ha J et al (2022) In situ identification of cellular drug targets in mammalian tissue. Cell 185:1793–1805. e17
Astre G, Moses E, Harel I (2022) The African turquoise killifish (Nothobranchius furzeri): biology and research applications. In: D’Angelo L, de Girolamo P (eds) Laboratory fish in biomedical research. Academic Press, pp 245–287
Coolen M, Labusch M, Mannioui A, Bally-Cuif L (2020) Mosaic heterochrony in neural progenitors sustains accelerated brain growth and neurogenesis in the juvenile Killifish N. furzeri. Curr Biol 30:736–745. e4
Lee S, Nam HG, Kim Y (2021) The core circadian component, Bmal1, is maintained in the pineal gland of old killifish brain. iScience 24:101905
Vanhunsel S, Bergmans S, Beckers A, Etienne I, Van Bergen T, De Groef L, Moons L (2022) The age factor in optic nerve regeneration: intrinsic and extrinsic barriers hinder successful recovery in the short-living killifish. Aging Cell 21:e13537
Acknowledgments
We thank Jeffrey Lange, Alice Accorsi, Dai Tsuchiya, Nancy Thomas, Seth Malloy, and Michael Frangello for meaningful discussion throughout this project. We also thank the Media Preparation Facility of the Stowers Institute for Medical Research for solutions preparation. We thank Robert Schnittker and Daniel Zamora for exceptional care of the killifish colony in the Sánchez Alvarado Lab. We thank Nicolas Denans for sharing the plasmid to generate the Scarlet transgenic killifish. This study is financially supported by the Stowers Institute for Medical Research.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Ortega Granillo, A., Deng, F., Wilson, H., Wang, Y., Sánchez Alvarado, A. (2023). Whole-Brain Clearing and Immunolabelling in the African Killifish Nothobranchius furzeri. In: Wang, W., Rohner, N., Wang, Y. (eds) Emerging Model Organisms. Neuromethods, vol 194. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-2875-1_4
Download citation
DOI: https://doi.org/10.1007/978-1-0716-2875-1_4
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-2874-4
Online ISBN: 978-1-0716-2875-1
eBook Packages: Springer Protocols