Abstract
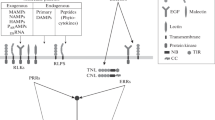
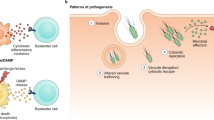
Detection of microbes by the host is essential to restrict microbial colonization, to clear pathogens, and to mount adapted defense reactions, and thus is the key function of the innate immune systems of plants and mammals. Here we provide an introduction into pathogen recognition by the innate immune system of both plants and animals. We will particularly focus on the concept of effector-triggered immunity, and similarities and differences in its function between plants and animals.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Janeway CA Jr (1989) Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb Symp Quant Biol 54(Pt 1):1–13
Medzhitov R, Janeway C Jr (2000) Innate immune recognition: mechanisms and pathways. Immunol Rev 173:89–97. https://doi.org/10.1034/j.1600-065x.2000.917309.x
Matzinger P (1994) Tolerance, danger, and the extended family. Annu Rev Immunol 12:991–1045. https://doi.org/10.1146/annurev.iy.12.040194.005015
Patel S (2018) Danger-associated molecular patterns (DAMPs): the derivatives and triggers of inflammation. Curr Allergy Asthma Rep 18(11):63. https://doi.org/10.1007/s11882-018-0817-3
Couto D, Zipfel C (2016) Regulation of pattern recognition receptor signalling in plants. Nat Rev Immunol 16(9):537–552. https://doi.org/10.1038/nri.2016.77
Jones JD, Vance RE, Dangl JL (2016) Intracellular innate immune surveillance devices in plants and animals. Science 354(6316):aaf6395. https://doi.org/10.1126/science.aaf6395
van Wersch S, Tian L, Hoy R, Li X (2020) Plant NLRs: the whistleblowers of plant immunity. Plant Commun 1(1):100016. https://doi.org/10.1016/j.xplc.2019.100016
Staskawicz BJ, Ausubel FM, Baker BJ, Ellis JG, Jones JD (1995) Molecular genetics of plant disease resistance. Science 268(5211):661–667. https://doi.org/10.1126/science.7732374
Jones JD, Dangl JL (2006) The plant immune system. Nature 444(7117):323–329. https://doi.org/10.1038/nature05286
Yuan M, Ngou BPM, Ding P, **n XF (2021) PTI-ETI crosstalk: an integrative view of plant immunity. Curr Opin Plant Biol 62:102030. https://doi.org/10.1016/j.pbi.2021.102030
Kufer TA, Creagh EM, Bryant CE (2019) Guardians of the cell: effector-triggered immunity steers mammalian immune defense. Trends Immunol 40(10):939–951. https://doi.org/10.1016/j.it.2019.08.001
Lopes Fischer N, Naseer N, Shin S, Brodsky IE (2020) Effector-triggered immunity and pathogen sensing in metazoans. Nat Microbiol 5(1):14–26. https://doi.org/10.1038/s41564-019-0623-2
Kufer TA, Fritz JH, Philpott DJ (2005) NACHT-LRR proteins (NLRs) in bacterial infection and immunity. Trends Microbiol 13(8):381–388. https://doi.org/10.1016/j.tim.2005.06.004
Dangl JL, Jones JD (2001) Plant pathogens and integrated defence responses to infection. Nature 411(6839):826–833. https://doi.org/10.1038/35081161
van der Hoorn RA, Kamoun S (2008) From guard to decoy: a new model for perception of plant pathogen effectors. Plant Cell 20(8):2009–2017. https://doi.org/10.1105/tpc.108.060194
Wani SH, Anand S, Singh B, Bohra A, Joshi R (2021) WRKY transcription factors and plant defense responses: latest discoveries and future prospects. Plant Cell Rep 40(7):1071–1085. https://doi.org/10.1007/s00299-021-02691-8
Saur IML, Panstruga R, Schulze-Lefert P (2021) NOD-like receptor-mediated plant immunity: from structure to cell death. Nat Rev Immunol 21(5):305–318. https://doi.org/10.1038/s41577-020-00473-z
Stuart LM, Paquette N, Boyer L (2013) Effector-triggered versus pattern-triggered immunity: how animals sense pathogens. Nat Rev Immunol 13(3):199–206. https://doi.org/10.1038/nri3398
Liston A, Masters SL (2017) Homeostasis-altering molecular processes as mechanisms of inflammasome activation. Nat Rev Immunol 17(3):208–214. https://doi.org/10.1038/nri.2016.151
Azimi T, Zamirnasta M, Sani MA, Soltan Dallal MM, Nasser A (2020) Molecular mechanisms of salmonella effector proteins: a comprehensive review. Infect Drug Resist 13:11–26. https://doi.org/10.2147/IDR.S230604
Aktories K (2011) Bacterial protein toxins that modify host regulatory GTPases. Nat Rev Microbiol 9(7):487–498. https://doi.org/10.1038/nrmicro2592
Berglund J, Gjondrekaj R, Verney E, Maupin-Furlow JA, Edelmann MJ (2020) Modification of the host ubiquitome by bacterial enzymes. Microbiol Res 235:126429. https://doi.org/10.1016/j.micres.2020.126429
Grishin AM, Beyrakhova KA, Cygler M (2015) Structural insight into effector proteins of Gram-negative bacterial pathogens that modulate the phosphoproteome of their host. Protein Sci 24(5):604–620. https://doi.org/10.1002/pro.2636
Brodsky IE, Medzhitov R (2009) Targeting of immune signalling networks by bacterial pathogens. Nat Cell Biol 11(5):521–526. https://doi.org/10.1038/ncb0509-521
Chambers KA, Scheck RA (2020) Bacterial virulence mediated by orthogonal post-translational modification. Nat Chem Biol 16(10):1043–1051. https://doi.org/10.1038/s41589-020-0638-2
Schroder K, Tschopp J (2010) The inflammasomes. Cell 140(6):821–832. https://doi.org/10.1016/j.cell.2010.01.040
Levinsohn JL, Newman ZL, Hellmich KA, Fattah R, Getz MA, Liu S, Sastalla I, Leppla SH, Moayeri M (2012) Anthrax lethal factor cleavage of Nlrp1 is required for activation of the inflammasome. PLoS Pathog 8(3):e1002638. https://doi.org/10.1371/journal.ppat.1002638
Chavarria-Smith J, Vance RE (2013) Direct proteolytic cleavage of NLRP1B is necessary and sufficient for inflammasome activation by anthrax lethal factor. PLoS Pathog 9(6):e1003452. https://doi.org/10.1371/journal.ppat.1003452
Chui AJ, Okondo MC, Rao SD, Gai K, Griswold AR, Johnson DC, Ball DP, Taabazuing CY, Orth EL, Vittimberga BA, Bachovchin DA (2019) N-terminal degradation activates the NLRP1B inflammasome. Science 364(6435):82–85. https://doi.org/10.1126/science.aau1208
Sandstrom A, Mitchell PS, Goers L, Mu EW, Lesser CF, Vance RE (2019) Functional degradation: a mechanism of NLRP1 inflammasome activation by diverse pathogen enzymes. Science 364(6435):eaau1330. https://doi.org/10.1126/science.aau1330
Pei G, Zyla J, He L, Moura-Alves P, Steinle H, Saikali P, Lozza L, Nieuwenhuizen N, Weiner J, Mollenkopf HJ, Ellwanger K, Arnold C, Duan M, Dagil Y, Pashenkov M, Boneca IG, Kufer TA, Dorhoi A, Kaufmann SH (2021) Cellular stress promotes NOD1/2-dependent inflammation via the endogenous metabolite sphingosine-1-phosphate. EMBO J 40(13):e106272. https://doi.org/10.15252/embj.2020106272
Jamilloux Y, Magnotti F, Belot A, Henry T (2018) The pyrin inflammasome: from sensing RhoA GTPases-inhibiting toxins to triggering autoinflammatory syndromes. Pathog Dis 76(3):fty020. https://doi.org/10.1093/femspd/fty020
Zhong Z, Liang S, Sanchez-Lopez E, He F, Shalapour S, Lin XJ, Wong J, Ding S, Seki E, Schnabl B, Hevener AL, Greenberg HB, Kisseleva T, Karin M (2018) New mitochondrial DNA synthesis enables NLRP3 inflammasome activation. Nature 560(7717):198–203. https://doi.org/10.1038/s41586-018-0372-z
Zhou R, Yazdi AS, Menu P, Tschopp J (2011) A role for mitochondria in NLRP3 inflammasome activation. Nature 469(7329):221–225. https://doi.org/10.1038/nature09663
Park YH, Wood G, Kastner DL, Chae JJ (2016) Pyrin inflammasome activation and RhoA signaling in the autoinflammatory diseases FMF and HIDS. Nat Immunol 17(8):914–921. https://doi.org/10.1038/ni.3457
Keestra AM, Winter MG, Auburger JJ, Frassle SP, Xavier MN, Winter SE, Kim A, Poon V, Ravesloot MM, Waldenmaier JF, Tsolis RM, Eigenheer RA, Baumler AJ (2013) Manipulation of small Rho GTPases is a pathogen-induced process detected by NOD1. Nature 496(7444):233–237. https://doi.org/10.1038/nature12025
Kufer TA, Kremmer E, Adam AC, Philpott DJ, Sansonetti PJ (2008) The pattern-recognition molecule Nod1 is localized at the plasma membrane at sites of bacterial interaction. Cell Microbiol 10(2):477–486. https://doi.org/10.1111/j.1462-5822.2007.01062.x
Bielig H, Lautz K, Braun PR, Menning M, Machuy N, Bruegmann C, Barisic S, Eisler SA, Andreel M, Zurek B, Kashkar H, Sansonetti PJ, Hausser A, Meyer TF, Kufer TA (2014) The cofilin phosphatase slingshot homolog 1 (SSH1) links NOD1 signaling to actin remodeling. PLoS Pathog 10(9):e1004351. https://doi.org/10.1371/journal.ppat.1004351
Legrand-Poels S, Kustermans G, Bex F, Kremmer E, Kufer TA, Piette J (2007) Modulation of Nod2-dependent NF-kappaB signaling by the actin cytoskeleton. J Cell Sci 120(Pt 7):1299–1310. https://doi.org/10.1242/jcs.03424
Rodrigues L, Graca RSF, Carneiro LAM (2018) Integrated stress responses to bacterial pathogenesis patterns. Front Immunol 9:1306. https://doi.org/10.3389/fimmu.2018.01306
West AP, Shadel GS, Ghosh S (2011) Mitochondria in innate immune responses. Nat Rev Immunol 11(6):389–402. https://doi.org/10.1038/nri2975
Maekawa T, Kufer TA, Schulze-Lefert P (2011) NLR functions in plant and animal immune systems: so far and yet so close. Nat Immunol 12(9):818–826. https://doi.org/10.1038/ni.2083
Urbach JM, Ausubel FM (2017) The NBS-LRR architectures of plant R-proteins and metazoan NLRs evolved in independent events. Proc Natl Acad Sci U S A 114(5):1063–1068. https://doi.org/10.1073/pnas.1619730114
Zhang L, Chen S, Ruan J, Wu J, Tong AB, Yin Q, Li Y, David L, Lu A, Wang WL, Marks C, Ouyang Q, Zhang X, Mao Y, Wu H (2015) Cryo-EM structure of the activated NAIP2-NLRC4 inflammasome reveals nucleated polymerization. Science 350(6259):404–409. https://doi.org/10.1126/science.aac5789
Wang J, Hu M, Wang J, Qi J, Han Z, Wang G, Qi Y, Wang HW, Zhou JM, Chai J (2019) Reconstitution and structure of a plant NLR resistosome conferring immunity. Science 364(6435):eaav5870. https://doi.org/10.1126/science.aav5870
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Kufer, T.A., Kaparakis-Liaskos, M. (2022). A Brief Introduction to Effector-Triggered Immunity. In: Kufer, T.A., Kaparakis-Liaskos, M. (eds) Effector-Triggered Immunity. Methods in Molecular Biology, vol 2523. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-2449-4_1
Download citation
DOI: https://doi.org/10.1007/978-1-0716-2449-4_1
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-2448-7
Online ISBN: 978-1-0716-2449-4
eBook Packages: Springer Protocols