Abstract
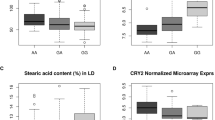
The associations of SNP in leptin gene (LEP g.2845A>T, LEP g.3996T>C, LEP g.3469T>C) and leptin receptor gene (LEPR c.232A>T, LEPR c.2856C>T, LEPR c.915C>T) with meat quality traits were studied in pigs of Ukrainian Large White breed (ULW). A computational analysis (SIFT, PolyPhen-2, and I-Mutant bioinformatic services) of the effect of mutations that cause SNPs (both for the selected SNP and for those missense SNPs, for which they can be potential linkage disequilibrium (LD) markers) on the structure and function of leptin and leptin receptor was carried out. The LEP c.3469T>C SNP (exon 3, rs45431504) was associated with the protein content in meat and moisture content in the back fat. The LEP g.2845A>T SNP (rs344615147), which is associated with the water-holding capacity of meat and moisture content in back fat, is located in the second intron and, as well as c.3469T>C SNP, can be presumably considered as an LD marker of the LEP rs701423985 SNP missense polymorphism. The LEPR c.232A>T SNP (exon 4, rs45435517) is a missense mutation, which causes the S52I amino acid substitution. The estimations obtained using SIFT and PolyPhen-2 indicate a significant effect of the rs45435517 on functional characteristics of leptin receptor. Indeed, an association of c.232A>T SNP with the thickness of back-fat was established in our work. At the same time, c.2856C>T SNP (exon 20, rs694660564, a synonymous substitution), which demonstrated in our work an association with the loss of moisture in the meat in ULW pigs, can be a potential LD marker and be in linkage disequilibrium with the LEPR rs1113972516, rs792804682, and rs1109261799 missense SNPs. SNP, for which the associations with meat quality indices were established in our work, can obviously be considered as potential markers for breeding ULW pigs and possibly other breeds aimed at obtaining genotypes with improved characteristics of meat quality.
Similar content being viewed by others
REFERENCES
Adzhubei, I., Jordan, D.M., and Sunyaev, S.R., Predicting functional effect of human missense mutations using PolyPhen-2, Curr. Protoc. Hum. Genet., 2013, vol. 7, no. 20. pp. 1–52. https://doi.org/10.1002/0471142905.hg0720s76
Balatsky, V., Bankovska, I., and Saienko, A., Association between leptin receptor gene polymorphism and quality of both meat and back fat in large white pigs of ukrainian breeding, Agric. Sci. Pract., 2016b, vol. 3, no. 2, pp. 42–48. https://doi.org/10.15407/agrisp3.02.042
Balatsky, V., Bankovska, I., Pena, R.N., et al., Polymorphisms of the porcine cathepsins, growth hormone-releasing hormone and leptin receptor genes and their association with meat quality traits in Ukrainian Large White breed, Mol. Biol. Rep., 2016a, vol. 43, pp. 517–526. https://doi.org/10.1007/s11033-016-3977-z
Balatsky, V., Oliinychenko, Y., Sarantseva, N., et al., Association of single nucleotide polymorphisms in leptin (LEP) and leptin receptor (LEPR) genes with backfat thickness and daily weight gain in Ukrainian Large White pigs, Livest. Sci., 2018, vol. 217, pp. 157–161. https://doi.org/10.1016/j.livsci.2018.09.015
Bauer, M., Babelova, A., Omelka, R., et al., Association of HinfI polymorphism in the leptin gene with production traits in White improved pig breed, Slovak J. Anim. Sci., 2006, vol. 39, pp. 119–122.
Bauer, M., Babelova, A., Omelka, R., et al., Effect of leptin and leptin receptor genes on meat production traits of Slovak Large White and Landrace pigs. Slovak J. Anim. Sci., 2009, vol. 42, pp. 49–53.
Biedermann, G. and Jatsch, C., Peschke, et al., Fattening and carcass performance and meat- and fat quality of Pietrain pigs of different MHS-genotype and sex. I. Fattening and carcass performance and meat quality, Archiv für Tierzucht, 2000, vol. 43, pp. 151–164.
Bižiene, R., Morkūnienė, K., Mišeikienė, R., et al., Effects of single nucleotide polymorphism markers on the carcass and fattening traits in different pig populations, J. Anim. Feed Sci., 2018, vol. 27, pp. 255–262. https://doi.org/10.22358/jafs/95020/2018
Boccard, R., Buchter, L., Casteels, M., et al., Procedures for measuring meat quality characteristics in beef production experiments, Livest. Prod. Sci., 1981, vol. 8, pp. 385–397. https://doi.org/10.1016/0301-6226(81)90061-0
Bonneau, M. and Lebret, B., Production systems and influence on eating quality of pork, Meat Sci., 2010, vol. 84, pp. 293–300. https://doi.org/10.1016/j.meatsci.2009.03.013
Breeding Strategies for Sustainable Management of Animal, FAO, Food and Agriculture Organization of the United Nation, 2014. http://www.fao.org/docrep/012/i1103e.pdf. Accessed October 28, 2019.
Capriotti, E., Fariselli, P., and Casadio, R.I., Mutant2.0: predicting stability changes upon mutation from the protein sequence or structure, Nucleic Acids Res., 2005, vol. 33, no. 2, pp. W306–W310. https://doi.org/10.1093/nar/gki375
Chmurzyńska, A., Maćkowski, M., Szydłowski, M., et al., Polymorphism of intronic microsatellites in the A-FABP and LEPR genes and its association with productive traits in the pig, J. Anim. Feed Sci., 2004, vol. 13, no. 4, pp. 615–624. https://doi.org/10.22358/jafs/67629/2004
Chorev, M. and Carmel, L., The function of introns, Front. Gene, 2012, vol. 3, p. 55. https://doi.org/10.3389/fgene.2012.00055
De Oliveira, P.J., Facioni, G.S.E., Sávio-Lopes, P., et al., Associations of leptin gene polymorphisms with production traits in pigs, J. Anim. Breed. Genet., 2006, vol. 123, pp. 378–383. https://doi.org/10.1111/j.1439-0388.2006.00611.x
De Oliveira, P.J., De Faria, D.A., Silva, P.V., et al., Association between leptin gene single nucleotide polymorphisms and carcass traits in pigs, Rev. Bras. Zootec., 2009, vol. 38, no. 2, pp. 271–276. https://doi.org/10.1590/S1516-35982009000200008
Dragos-Wendrich, M., Stratil, A., Hojny, J., et al., Linkage and QTL map** for Sus scrofa chromosome 18, J. Anim. Breed. Genet., 2003, vol. 120, pp. 138–143.
European Convention for the Protection of Vertebrate Animals used for Experimental and Other Scientific Purposes, Strasbourg, 18.III.1986. Retrieved from http:// conventions.coe.int/treaty/en/treaties/html/123.htm. Accessed March 12, 2019.
Galve, A., Burgos, C., Silió, L., et al., The effects of leptin receptor (LEPR) and melanocortin-4 receptor (MC4R) polymorphisms on fat content, fat distribution and fat composition in a Duroc × Landrace/Large White cross, Livest. Sci., 2012, vol. 145, pp. 145–152. https://doi.org/10.1016/j.livsci.2012.01.010
Getmantseva, L., Leonova, M., Usatov, A., et al., The single and combined effect of MC4R and GH genes on productive traits of pigs, Am. J. Agric. Biol. Sci., 2017, vol. 12, pp. 28–32. https://doi.org/28-32.10.3844/ajabssp.2017.28.32
Henriquez-Rodriguez, E., Bosch, L., Tor, M., et al., The effect of SCD and LEPR genetic polymorphisms on fat content and composition is maintained throughout fattening in Duroc pigs, Meat Sci., 2016, vol. 121, pp. 33–39. https://doi.org/10.1016/j.meatsci.2016.05.012
Hermesch, S., Li, L., Doeschl-Wilson, A.B., et al., Selection for productivity and robustness traits in pigs, Anim. Prod. Sci., 2015, vol. 55, no. 11, pp. 1437–1447. https://doi.org/10.1071/AN15275
Hirose, K., Ito, T., Fukawa, K., et al., Evaluation of effects of multiple candidate genes (LEP, LEPR, MC4R, PIK3C3, and VRTN) on production traits in Duroc pigs, Anim. Sci. J., 2014, vol. 85, pp. 198–206. https://doi.org/10.1111/asj.12134
Jiang, Z.H. and Gibson, J.P., Genetic polymorphisms in the leptin gene and their association with fatness in four pig breeds, Mamm. Genome, 1999, vol. 10, pp. 191–193.
Kennes, Y.M., Murphy, B.D., Pothier, F., et al., Characterization of swine leptin (LEP) polymorphisms and their association with production traits, Anim. Genet., 2001, vol. 32, pp. 215–218. https://doi.org/10.1046/j.1365-2052.2001.00768.x
Kim, J.-M., Park, J.-E., Lee, S.-W., et al., Association of polymorphisms in the 5′ regulatory region of LEPR gene with meat quality traits in Berkshire pigs, Anim. Genet., 2017, vol. 48, no. 6, pp. 723–724. https://doi.org/10.1111/age.12588
Kim, J.A., Cho, E.S., Jeong, Y.D., et al., The effects of breed and gender on meat quality of Duroc, Pietrain, and their crossbred, J. Anim. Sci. Technol., 2020, vol. 62, pp. 409–419. https://doi.org/10.5187/jast.2020.62.3.409
Kramarenko, A.S., Ignatenko, Zh.V., Lugovoy, S.I., et al., Effect of parity number, year and season farrowing on reproductive performance in Large White pigs, Ukr. J. Ecol., 2020, vol. 10, pp. 307–312. https://doi.org/10. 15421/2020_48
Li, X., Kim, S.W., Choi, J.S., et al., Investigation of porcine FABP3 and LEPR gene polymorphisms and mRNA expression for variation in intramuscular fat content, Anim. Genet., 2005, vol. 36, pp. 135–137. https://doi.org/10.1111/j.1365-2052.2005.01247.x
Li, X., Kim, S.W., Choi, J.S., et al., Investigation of porcine FABP3 and LEPR gene polymorphisms and mRNA expression for variation in intramuscular fat content, Mol. Biol. Rep., 2010, vol. 37, pp. 3931–3939. https://doi.org/10.1007/s11033-010-0050-1
Malek, M., Dekkers, J.C., Lee, H.K., et al., A molecular genome scan analysis to identify chromosomal regions influencing economic traits in the pig. II. Meat and muscle composition, Mamm. Genome, 2001, vol. 12, pp. 637–645. https://doi.org/10.1007/s003350020019
Mankowska, M., Szydlowski, M., Salamon, S., et al., Novel polymorphisms in porcine 3’UTR of the leptin gene, including a rare variant within target sequence for MIR-9 gene in Duroc breed, not associated with production traits, Anim. Biotechnol., 2015, vol. 26, no. 2, pp. 156–163. https://doi.org/10.1080/10495398.2014.958612
Matoulkova, E., Michalova, E., Vojtesek, B., et al., The role of the 3’ untranslated region in post-transcriptional regulation of protein expression in mammalian cells, RNA Biol., 2012, vol. 9, no. 5, pp. 563–576. https://doi.org/10.4161/rna.20231
Matousek, V., Kernerova, N., Hysplerová, K., et al., Carcass traits and meat quality of Prestice Black-Pied pig breed, Asian-Australas. J. Anim. Sci., 2016, vol. 29, pp. 1181–1187. https://doi.org/10.5713/ajas.15.0659
Munoz, G., Ovilo, C., Silió, L., et al., Single and joint-population analyses of two experimental pig crosses to confirm quantitative trait loci on Sus scrofa chromosome 6 and leptin receptor effects on fatness and growth traits, J. Anim. Sci., 2009, vol. 87, pp. 459–468. https://doi.org/10.2527/jas.2008-1127
Ng, P.C. and Henikoff, S., SIFT: predicting amino acid changes that affect protein function, Nucleic Acids Res., 2003, vol. 31, no. 13, pp. 3812–3814. https://doi.org/10.1093/nar/gkg509
Official Methods of Analysis, Arlington: Assoc. Off. Anal. Chem., 1990, 15th ed.
Oleinychenko, E.K., Bankovskaia, Y.B., Balatsky, V.N., et al., Henetychnyi ta asotsiatyvnyi analiz odnonukleotydnykh polimorfizmiv v henakh leptynu I katepsynu F svynei. Naukovyi visnyk Natsionalnoho universytetu bioresursiv i pryrodokorystuvannia Ukrainy, Ser.: Tekhnolohiia vyrobnytstva i pererobky produktsii tvarynnytstva, 2018a, vol. 289, pp. 38–50.
Oleinychenko, E.K., Sarantseva, N.K., Vovk, V.A., et al., Vlyianye polymorfyzmov henov leptyna y retseptora leptyna na produktyvnûe kachestva svynei krupnoi beloi porodû, Svynarstvo: Mizhvid. Temat. Nauk, Zb., 2018b, vol. 71, pp. 83–92.
Óvilo, C., Fernández, A., Noguera, J.L., et al., Fine map** of porcine chromosome 6 QTL and LEPR effects on body composition in multiple generations of an Iberian by Landrace intercross, Genet. Res., 2005, vol. 85, pp. 57–67. https://doi.org/10.1017/s0016672305007330
Pant, S.D., Karlskov-Mortensen, P., Jacobsen, M.J., et al., Comparative analyses of QTLs influencing obesity and metabolic phenotypes in pigs and humans, PLoS One, 2015, vol. 10, no. 9, p. e0137356.https://doi.org/10.1371/journal.pone.0137356
Peakall, R. and Smouse, P.E., GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update, Bioinformatics, 2012, vol. 28, pp. 2537–2539.
Pena, R.N., Ros-Freixedes, R., Tor, M., et al., Genetic marker discovery in complex traits: a field example on fat content and composition in pigs, Int. J. Mol. Sci., 2016, vol. 17, p. 2100. https://doi.org/10.3390/ijms17122100
Ros-Freixedes, R., Gol, S., Pena, R.N., Tor, M., et al., Genome-wide association study singles out SCD and LEPR as the two main loci influencing intramuscular fat content and fatty acid Ccmposition in Duroc pigs, PLoS One, 2016, vol. 11, pp. 1–18. https://doi.org/10.1371/journal.pone.0152496
Szydlowski, M., Stachowiak, M., Mackowski, M., et al., No major effect of the leptin gene polymorphism on porcine production traits, J. Anim. Breed. Genet., 2004, vol. 121, pp. 149–155. https://doi.org/10.1111/j.1439-0388.2004.00453.x
Trakovicka, A., Moravčíková, N., Kukučková, V., et al., The associations of lepr and H-FABP gene polymorphisms with carcass traits in pigs, Acta Argiculturae Slovenica, 2016, vol. 5, pp. 189–194.
Uemoto, Y., Kikuchi, T., Nakano, H., et al., Effects of porcine leptin receptor gene polymorphisms on backfat thickness, fat area ratios by image analysis, and serum leptin concentrations in a Duroc purebred population, Anim. Sci. J., 2012, vol. 83, pp. 375–385. https://doi.org/10.1111/j.1740-0929.2011.00963.x
Vega, R.S.A., Castillo, R.M.C., Barientos, N.N.B., et al., Leptin (T3469C) and estrogen receptor (T1665G) gene polymorphisms and their associations to backfat thickness and reproductive traits of Large White pigs (Sus scrofa L.), Philippine J. Sci., 2018, vol. 147, no. 2, pp. 293–300.
Villalba, D., Tor, M., Vidal, O., et al., An age-dependent association between a leptin C3469T single nucleotide polymorphism and intramuscular fat content in pigs, Livestock Sci., 2009, vol. 121, pp. 335–338. https://doi.org/10.1016/j.livsci.2008.06.008
Zhang, C.Y., Wang, Z., Bruce, H.L., et al., Associations between single nucleotide polymorphisms in 33 candidate genes and meat quality traits in commercial pigs, Anim. Genet., 2014, vol. 45, pp. 508–516. https://doi.org/10.1111/age.12155
Funding
This work was supported by the Institute of Pig Breeding and Agroindustrial Production, National Academy of Agrarian Sciences of Ukraine (research no. 0111U005008), the University of the West of England, Bristol and EU COST Action IPEMA (grant no. CA15215).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest. The authors declare that they have no conflicts of interest.
Statement on the welfare of animals. All procedures related to the treatment of animals were in accordance with the European Convention for the Protection of Vertebrate Animals Used for Experimental and Other Scientific Purposes (European Convention, 1986).
Additional information
Translated by A. Barkhash
About this article
Cite this article
Balatsky, V.N., Oliinychenko, Y.K., Saienko, A.M. et al. Associations of Polymorphisms in Leptin and Leptin Receptor Genes with Meat Quality in Pigs of the Ukrainian Large White Breed. Cytol. Genet. 56, 513–525 (2022). https://doi.org/10.3103/S0095452722060020
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.3103/S0095452722060020