Abstract
Molecular mechanisms underlying bipolar affective disorders are unknown. Difficulties arise from genetic and phenotypic heterogeneity of patients and the lack of animal models. Thus, we focused on only one patient (n = 1) with an extreme form of rapid cycling. Ribonucleic acid (RNA) from peripheral blood mononuclear cells (PBMC) was analyzed in a three-tiered approach under widely standardized conditions. Firstly, RNA was extracted from PBMC of eight blood samples, obtained on two consecutive days within one particular episode, including two different consecutive depressive and two different consecutive manic episodes, and submitted to (1) screening by microarray hybridizations, followed by (2) detailed bioinformatic analysis, and (3) confirmation of episode-specific regulation of genes by quantitative real-time polymerase chain reaction (qRT-PCR). Secondly, results were validated in additional blood samples obtained one to two years later. Among gene transcripts elevated in depressed episodes were prostaglandin D synthetase (PTGDS) and prostaglandin D2 11-ketoreductase (AKR1C3), both involved in hibernation. We hypothesized them to account for some of the rapid cycling symptoms. A subsequent treatment approach over 5 months applying the cyclooxygenase inhibitor celecoxib (2 × 200 mg daily) resulted in reduced severity rating of both depressed and manic episodes. This case suggests that rapid cycling is a systemic disease, resembling hibernation, with prostaglandins playing a mediator role.
Similar content being viewed by others
Introduction
Rapid cycling syndrome is a bipolar affective disorder, amounting to 10% to 30% of the bipolar population. It is characterized by at least four episodes per year and rapid shifts between cycles. Patients with bipolar affective disorder, as well as patients with rapid cycling syndrome, typically experience their first major mood episode during adolescence (1–5).
Recently, gene expression data from post mortem brains of bipolar patients were compared with those of healthy controls in two independent studies (6,7). While post mortem approaches certainly cannot reveal cyclic changes of gene expression, these studies also failed to yield a single overlap** candidate gene for bipolar disease. Moreover, the lack of an adequate animal model for bipolar disorder demands novel experimental approaches.
We hypothesized that cycling alterations of brain functions in bipolar disease are reflected by systemic physiological changes that have a molecular genetic basis. If true, it should be possible to obtain molecular signatures of manic and depressed states even outside the brain, such as in peripheral blood mononuclear cells (PBMC). While not disease causing, such gene expression changes in PBMC may shed light on similar cyclic alterations in brain.
To study quantitative peripheral gene expression, we specifically refrained from comparing larger groups of bipolar patients (who are genetically heterogeneous and differ in baseline gene expression profiles), and aimed instead at monitoring the gene expression in one individual, serving always as her own control, at recurrent stages of the disease.
Case Report
The female patient, born in 1945, had no prior medical illness and no evidence of neuropsychiatric illnesses in her family. In 1991, she became ill with rapid cycling syndrome and kept a diary over her illness, used to reconstruct 108 cycles over a 16-year period. The time series suggests complex rhythms in periodicity with mean total cycle lengths of 53 ± 21 d, switching within hours between manic (mean 28 ± 14 d) and depressed (mean 26 ± 14 d) episodes without normal intervals (Supplementary Figure 1). Results of affective rating scales obtained repeatedly during depressed and manic episodes, together with psychopathology, neuropsychological test results, appearance, autonomic, and physical signs are summarized in Table 1.
In addition to typical affective symptoms, the patient has physical and cognitive signs recurring in an episode-specific manner. In the first 2 to 3 d of a manic episode, she is sleepless and restless; in the following d, she sleeps 3 to 4 h per night. The 2 to 3 d before the end of manic episodes, she notes a ‘normalization of sleep’ with non-interrupted sleep of regular 8 h duration. The patient eats and drinks excessively during manic episodes, leading to alternating weight changes (up to 5 kg) between episodes and hyperhydration, resulting in significant shifts of hematocrit and hemoglobin concentrations. Three d after the onset of manic episodes, the patient regularly develops edema in her lower extremities that recover immediately after onset of depression. Only during manic episodes does she become susceptible to seasonal allergies (hay fever). This allergic response is rarely observed during depressed episodes. Witnesses describe a change of her voice in the last 2 to 3 d of manic episodes to raspy and less melodious. At the end of depressed episodes, her voice becomes more cheerful and richer in tonal inflections. The patient is not aware of these changes. Because there was an episode-specific susceptibility to allergens, lymphocyte subpopulations were studied by fluorescence-activated cell sorting in different episodes. Subtle shifts between CD4-helper and CD8-suppressor cells were noted (Supplementary Figure 2).
The cyclic pattern of the patient’s affective disorder has had a poor response to pharmacologic treatment over the past 16 years, such as lithium, venlafaxine, chlorprothixene, citalopram, paroxetine, carbamazepine, valproic acid, trimipramine, lamotrigine, olanzapine, or flupentixol, and no response to psychotherapy and hypnosis. Antipsychotic medication, such as flupentixol, somewhat reduced the severity of symptoms but not the cyclic behavior of the disorder. During the time of all analyses presented here, the patient was on continuous lamotrigine medication (400 mg), resulting in comparable serum levels of 4.1–8.9 µg/mL upon repeated controls (therapeutic range: 2–10 µg/mL). No other medication was allowed 2 wks before or during the time of each testing, or before and during the treatment approach, reported here, using the cyclooxygenase inhibitor celecoxib (Celebrex, Pfizer, Karlsruhe, Germany, 2 × 200 mg daily per os).
Materials and Methods
Strategy of Episode-Dependent Gene Shift Detection
A three-tiered approach was used to identify candidate genes that are expressed in an episode-specific fashion. In the first step, eight blood samples for PBMC isolation (see below) were collected (always at 8:00 a.m. after overnight fasting conditions) in the approximate middle of two different consecutive depressed and manic episodes on two consecutive days each. All sample collection was done well outside the hay fever season and in complete absence of allergic symptoms. The screening identified genes that showed at least two-fold differences in expression in manic compared with depressed episodes, and vice versa. After screening by microarrays, the data set was submitted to two bioinformatic processing steps (see below). The data were normalized and analyzed to identify and exclude genes that differed between the two consecutive days within a particular episode (arbitrary daily variation). Further genes were excluded that were differentially expressed within the two depressed or within the two manic episodes (arbitrary monthly variation). The expression pattern of the remaining depressed and manic episode genes was subsequently compared. Using this approach, several genes were identified that showed high expression in depressed and low in manic episodes, and vice versa. Because the patient never had periods where she was free of symptoms, samples could not be obtained within an euthymic episode.
Isolation of Peripheral Blood Mononuclear Cells (PBMC)
PBMC were collected applying the standard Ficoll-Paque Plus isolation procedure (Amersham Biosciences, Freiburg, Germany). RNA was prepared using Trizol and Qiagen RNAeasy columns (Qiagen, Hilden, Germany). The RNA samples were used to synthesize cDNAs (SuperScriptIII, Invitrogen, Karlsruhe, Germany).
DNA Microarray Analysis
Transcriptome analysis was performed using the GeneChip Human Genome U133 Plus 2.0 (Affymetrix, Santa Clara, CA, USA) (8) according to the published protocols (9). All cDNAs used for microarrays were one-round amplified. GeneChip data were analyzed using the software GCOS version 1.2 (Affymetrix). Detailed data analysis was performed with R open-source software, and the open-source Bioconductor platform.
Quantitative Real-Time Reverse Transcriptase Polymerase Chain Reaction (qRT-PCR)
Genes found to be differentially regulated by DNA microarray analysis were subsequently validated independently using qRT-PCR. Cyclic changes in the expression of these genes were confirmed further on PBMC of additional blood samples obtained up to two years after the initial screen. qRT-PCR was performed with the aid of SYBR Green detection on Applied Biosystems 7500 Fast System. CT (cycle threshold) values were standardized to CT values of GAPDH. Primers are listed in Supplementary Table 1.
Statistical Analyses
All numerical results are presented as mean ± SD in the text and mean ± SEM in the Figures. Statistical analyses (10,11) and Fast Fourier Transformations (12) were performed as published using MATLAB R2007a software. Nonparametric independent Mann-Whitney U test (two-tailed) and Student t test (two-tailed) were calculated using SPSS 16.0 for Windows.
All supplementary materials are available online at molmed.org .
Results
A three-tiered approach was employed to identify genes that were regulated in an episode-specific fashion (Figure 1A). We grouped the genes in biological categories and present their mean expression pattern over six to ten separate time points during either depression or mania (Figure 1B–E).
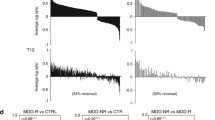
(A) Strategy of episode-dependent gene shift detection using microchip analysis. A three-step strategy was used to identify candidate genes that are expressed in an episode-specific fashion: Eight blood samples were collected (always at 8:00 a.m.) in two consecutive depressed and manic episodes on two consecutive days each. After screening by microarrays, the data set was submitted to two bioinformatic processing steps. Genes were subtracted that differed between the two consecutive days within a particular episode (arbitrary daily variation). Further genes were excluded that were differentially expressed within the two depressed or within the two manic episodes (arbitrary monthly variation). The expression pattern of the remaining depressed and manic episode genes was subsequently compared. (B–E) Episode-specific PBMC gene expression involves different groups of genes. Genes found to be differentially expressed by microchip screening were confirmed by qRT-PCR in all samples. Moreover, blood sampling was extended beyond the initial screening period, and regulated genes were again validated more than one year later. Genes were grouped (B–E) according to biological categories. Each bar represents mean ± SEM of 6 to 12 determinations from cDNAs obtained from PBMC on different days in independent manic (n = 6; open bars) and depressed (n = 5; filled bars) episodes. The chart for hemoglobins contains an insert with hemoglobin (protein) concentrations in whole blood (n = 7 samples of independent manic and depressed episodes, respectively). Mean values were compared using independent Student t test (two-tailed).
Notably, the genes involved in prostaglandin metabolism, PTGDS (lipocalin-type prostaglandin D synthetase), and AKR1C3 (prostaglandin D2 11-ketoreductase), showed higher expression in depressed episodes (Figure 1B). We also identified two neurodevelopmental genes that revealed opposing gene expression: NRG1 (neuregulin-1) was expressed higher in manic, SPON2 (spondin-2) in depressed episodes (Figure 1C). Several other genes, involved in regulation of the immune system, were episode-specifically expressed. These include GZMA (granzyme A) and KLRD1 (killer cell lectin-like receptor subfamily D, member 1/CD94) with higher expression in depressed as compared with manic episodes (Figure 1D). Hemoglobin genes A and B (HBB and HBA) were higher in manic, compared with depressed episodes. This contrasts the hemoglobin (protein) concentration in whole blood (insert) showing opposite regulation (Figure 1E).
As discussed below, the most intriguing finding was the association of prostaglandin-synthesizing genes PTGDS (lipocalin-type prostaglandin D synthetase) and AKR1C3 (prostaglandin D2 11-ketoreductase) with rapid cycling. We undertook a clinical experiment to explore the relevance of this observation and to distinguish between a disease marker and mediator. We offered to the patient a treatment approach applying (off-label) the cyclooxygenase inhibitor celecoxib (Celebrex, Pfizer, 2 × 200 mg daily oral).
Treatment with celecoxib was started with 100 mg and increased by 100 mg daily to reach the final dose of 400 mg (2 × 200 mg daily) at d 4. This dose has been continued for more than 5 months and is well tolerated. Figure 2 illustrates the clinical course before and during celecoxib, including psychopathology ratings that revealed a significant improvement of depressed as well as of manic symptoms.
Clinical course of psychopathology ratings before and during treatment with the cyclooxygenase inhibitor celecoxib. The course of the Hamilton Depression Rating Scale (HAM-D) scores, the Young Mania Rating Scale (YMRS) scores, and the Positive and Negative Syndrome Scale (PANSS) scores, presented in line graphs, show a pronounced cycling pattern. Day 0 denotes start of celecoxib intake. The magnitude of cyclic changes appears to gradually decrease during treatment. Bar graphs give scores (mean ± SEM) of n = 6 depressed and n = 6 manic episodes (two before and four after onset of treatment each). HAM-D score in depressed and YMRS score in manic episodes decrease significantly upon treatment. Mean values are compared using independent Student t test (two-tailed). M = manic episode, D = depressed episode.
Discussion
In our first molecular-genetic approach to alternating gene expression in bipolar disorders, we used PBMC of a woman with 16-year history of an extreme form of rapid cycling, and obtained an episode-specific gene expression profile on an identical genetic background. Our strategy was set up to minimize the risk of identifying false positive genes, due to daily or monthly variations in gene expression that are disorder-unrelated. The gene expression differences among episodes are small but significant, and would not have been recognized in a pool of patients or in comparison with healthy controls, due to different genetic background and modifiers of gene expression. In principle, such an approach could be employed as a screening strategy for genes in any condition with temporal periodic behavior, such as sleep or seasonal phenomena/disorders.
Follow-up studies on larger numbers of bipolar patients will have to follow, to confirm the general disease-relevance of the identified gene expression shifts. Specifically, patients with a more typical age of onset and course of bipolar disease will have to be screened. An age of onset of 46 years and cycling over decades without euthymic episodes, as in our patient, clearly is an exception (3–5).
The patient’s impairments comprise psychopathological symptoms in combination with physical signs and symptoms, including the immune system, and a variety of cognitive domains evident upon neuropsychological testing. Accordingly, episode-specific gene expression involved different biological systems, such as blood, metabolism, immune functions, as well as neuronal genes, confirming rapid cycling as a systemic disorder. Like all association studies, we can make no claim whether or not a particular gene expression shift is a cause or consequence of rapid cycling, and whether similar gene expression changes occur in the brain. Moreover, it is currently unknown whether some of the observed alterations in gene expression partly reflect the shifts in immune cell subsets observed here.
Lamotrigine has been shown to alter expression of certain genes, such as GABA-A receptor β3 subunit in rat hippocampal cells (13). During the entire observation period (analysis and experimental treatment) reported in this paper, however, the patient was always on the same dose of lamotrigine (episode-independent), making an influence of this pharmacological treatment on the alternating gene expression shown here very unlikely. No other medication was used.
Some regulated genes identified in PBMC are known to be expressed in the nervous system. For example, NRG1 is a neuronal growth factor regulating differentiation, synaptogenesis, and myelination of the nervous system (14). NRG1 also is expressed in activated monocytes (15). Similarly, SPON2 (spondin, mindin) originally was characterized in zebrafish as a protein involved in outgrowth of hippocampal neurons (16). It is expressed abundantly in lymphoid tissue and involved in inflammation (17).
Granzymes A and B (GZMA and GZMB) and several natural killer cell receptors were higher in depressed episodes. We speculate that the observed episode-specific gene expression contributes to the pathogenesis seen in our patient, including her allergic diathesis. In contrast, the elevated globin gene expression in manic episodes found here may reflect a physiological counter-regulation to the relative increase in extracellular fluid (and decrease in hemoglobin level) due to massive drinking.
Interestingly, two genes were identified that are essential for prostaglandin synthesis, PTGDS and AKR1C3. PTGDS is preferentially expressed in the central nervous system and mediates synthesis of PGD2 from PGH2 (the cyclooxygenase-mediated product of arachidonic acid), and AKR1C3 mediates synthesis of PGF2α from PGD2. PGF2α is a PPARγ antagonist, in contrast to two other PGD2 metabolites that are spontaneously converted from PGJ2: A12-PGJ2 and 15-deoxy-∆12,14-PGJ2 that are PPARγ agonists. Prostaglandin synthesis plays a pivotal role in metabolic homeostasis, sleep regulation, adipogenesis, allergic response, and inflammation (18–22). Altered levels of prostaglandins have been detected in different body fluids in patients with major affective disorders (23–28). Also pharmacological evidence (29–31) suggests that altered prostaglandin metabolism may lead to mood disorders. All this is consistent with episode-specific differential gene expression of PTGDS and AKR1C3 as revealed in the present case.
Alterations in inflammatory markers in serum of bipolar patients, such as cytokines, c-reactive protein and components of the complement system, have been reported previously (32–38). There might well be an interaction between the prostaglandin system found to be episodically regulated here, in the described case, and these inflammatory molecules, also in the CNS (35,39,40).
Intriguingly, altered expression of PTGDS marks the hibernation cycle, and accumulation of prostaglandins during hibernation season has been described in hibernating animals (18,22). Based on our molecular data, we speculate that the rapid cycling syndrome in humans may reflect an evolutionary ancient behavioral program resembling the hibernation cycle (with periodic eating, high psychomotor activity and nesting behavior, alternating with episodes of rest and sleep) (41,42) that becomes pathologically re-activated by unknown triggers, thereby creating rapid cycling.
The gene products involved in prostaglandin metabolism might give clues to potential treatment targets, but to confirm their general relevance in bipolar disease, more patients will have to be examined for cycling-associated alterations in prostaglandin gene expression. Nevertheless, the positive result of the clinical experiment with celecoxib in our patient, showing considerable attenuation of both depressed and manic rating scores, supports a mediator role of prostaglandins in rapid cycling. We are fully aware of the limitation of a case report in comparison to a clinical trial. However, we note that the 16-year clinical history of our patient has never shown benefits from any of the many pharmacological treatments, suggesting that a potential placebo effect in our experiment would be minor.
Supporting our findings, a recent proof-of-concept trial in Germany including patients with major monopolar depression found beneficial effects on mood upon 6-wk add-on treatment with the cyclooxygenase-2 inhibitor celecoxib. In contrast to our gene expression-based “hibernation hypothesis of bipolar disease,” this trial was based on the hypothesis that inflammatory processes might be involved in the pathogenesis of depression (43). Following the same inflammation hypothesis of depression, another small 6-wk study was performed in the United States of America, exploring the effect of celecoxib as adjunctive agent in bipolar depression, also with promising results (44). Bringing hibernation and inflammation together, it is intriguing to speculate that rapid cycling bipolar disorders are characterized by episodic self-limiting inflammatory processes that, unlike other inflammatory conditions in the brain, do not lead to overt neurodegeneration.
Disclosure/Conflicts of Interest
It is herewith declared that none of the authors have any conflicts of interest in publishing our data.
References
Dunner DL, Fieve RR. (1974) Clinical factors in lithium-carbonate prophylaxis failure. Arch. Gen. Psychiatry 30:229–33.
Muller-Oerlinghausen B, Berghofer A, Bauer M. (2002) Bipolar disorder. Lancet 359:241–7.
Coryell W. (2005) Rapid cycling bipolar disorder: clinical characteristics and treatment options. CNS Drugs 19:557–69.
Papadimitriou GN, Calabrese JR, Dikeos DG, Christodoulou GN. (2005) Rapid cycling bipolar disorder: biology and pathogenesis. Int. J. Neuropsychopharmacol. 8:281–92.
Schneck CD, et al. (2004) Phenomenology of rapid-cycling bipolar disorder: data from the first 500 participants in the Systematic Treatment Enhancement Program. Am. J. Psychiatry 161:1902–8.
Nakatani N, et al. (2006) Genome-wide expression analysis detects eight genes with robust alterations specific to bipolar I disorder: relevance to neuronal network perturbation. Hum. Mol. Genet. 15:1949–62.
Ryan MM, et al. (2006) Gene expression analysis of bipolar disorder reveals downregulation of the ubiquitin cycle and alterations in synaptic genes. Mol. Psychiatry 11:965–78.
Shi LM, et al. (2006) The MicroArray Quality Control (MAQC) project shows inter- and intra-platform reproducibility of gene expression measurements. Nat. Biotechnol. 24:1151–61.
Rossner MJ, et al. (2006) Global transcriptome analysis of genetically identified neurons in the adult cortex. J. Neurosci. 26:9956–66.
Hollander M, Wolfe DA. (1999) Nonparametric Statistical Methods. J. Wiley, New York.
Stuart A, Ord JK, Arnold S. (1999) Kendall’s Advanced Theory of Statistics. (Vol. 2A). Hodder Arnold, London.
Duhamel P, Vetterli M. (1990) Fast Fourier transforms: a tutorial review and state of the art. Signal Processing 19:259–99.
Wang JF, Sun X, Chen B, Young LT. (2002) Lamotrigine increases gene expression of GABA-A receptor beta3 subunit in primary cultured rat hippocampus cells. Neuropsychopharmacology 26:415–21.
Role LW, Talmage DA. (2007) Neurobiology: new order for thought disorders. Nature. 448:263–5.
Mograbi B, et al. (1997) Human monocytes express amphiregulin and heregulin growth factors upon activation. Eur. Cytokine Netw. 8:73–81.
Feinstein Y, et al. (1999) F-spondin and mindin: two structurally and functionally related genes expressed in the hippocampus that promote outgrowth of embryonic hippocampal neurons. Development 126:3637–48.
Jia W, Li H, He YW. (2005) The extracellular matrix protein mindin serves as an integrin ligand and is critical for inflammatory cell recruitment. Blood 106:3854–9.
O’Hara BF, et al. (1999) Gene expression in the brain across the hibernation cycle. J. Neurosci. 19:3781–90.
Hayaishi O. (2002) Functional genomics of sleep and circadian rhythm: Invited review: Molecular genetic studies on sleep-wake regulation, with special emphasis on the prostaglandin D2 system. J. Appl. Physiol. 92:863–8.
Qu WM, et al. (2006) Lipocalin-type prostaglandin D synthase produces prostaglandin D2 involved in regulation of physiological sleep. Proc. Natl. Acad. Sci. U. S. A. 103:17949–54.
Ram A, et al. (1997) CSF levels of prostaglandins, especially the level of prostaglandin D2, are correlated with increasing propensity towards sleep in rats. Brain Res. 751:81–9.
Takahata R, et al. (1996) Seasonal variation in levels of prostaglandins D2, E2 and F2(alpha) in the brain of a mammalian hibernator, the Asian chipmunk. Prostaglandins Leukot. Essent. Fatty Acids 54:77–81.
Bennett CN, Horrobin DF. (2000) Gene targets related to phospholipid and fatty acid metabolism in schizophrenia and other psychiatric disorders: an update. Prostaglandins Leukot. Essent. Fatty Acids 63:47–59.
Calabrese JR, Gulledge AD. (1984) Prostaglandin E2 and depression. Biol. Psychiatry 19:1269–71.
Horrobin DF, Manku MS. (1980) Possible role of prostaglandin E1 in the affective disorders and in alcoholism. Br. Med. J. 280:1363–6.
Lieb J, Karmali R, Horrobin D. (1983) Elevated levels of prostaglandin E2 and thromboxane B2 in depression. Prostaglandins Leukot. Med. 10:361–7.
Lowinger P. (1989) Prostaglandins and organic affective syndrome. Am. J. Psychiatry 146:1646–7.
Ohishi K, Ueno R, Nishino S, Sakai T, Hayaishi O. (1988) Increased level of salivary prostaglandins in patients with major depression. Biol. Psychiatry 23:326–34.
Ansell D, Belch JJ, Forbes CD. (1986) Depression and prostacyclin infusion. Lancet 2:509.
Bishop LC, Bisset AD, Benson JI. (1987) Mania and indomethacin. J. Clin. Psychopharmacol. 7:203–4.
Lloyd DB. (1992) Depression on withdrawal of indomethacin. Br. J. Rheumatol. 31:211.
Cunha AB, et al. (2008) Investigation of serum high-sensitive C-reactive protein levels across all mood states in bipolar disorder. Eur. Arch. Psychiatry Clin. Neurosci. 2008, Feb 23. [Epub ahead of print].
Dickerson F, Stallings C, Origoni A, Boronow J, Yolken R. (2007) Elevated serum levels of C-reactive protein are associated with mania symptoms in outpatients with bipolar disorder. Prog. Neuropsychopharmacol. Biol. Psychiatry 31:952–5.
Hornig M, Goodman DB, Kamoun M, Amsterdam JD. (1998) Positive and negative acute phase proteins in affective subtypes. J. Affect. Disord. 49: 9–18.
Kim YK, Jung HG, Myint AM, Kim H, Park SH. (2007) Imbalance between pro-inflammatory and anti-inflammatory cytokines in bipolar disorder. J. Affect. Disord. 104:91–5.
O’Brien SM, Scully P, Scott LV, Dinan TG. (2006) Cytokine profiles in bipolar affective disorder: focus on acutely ill patients. J. Affect. Disord. 90: 263–7.
Ortiz-Dominguez A, et al. (2007) Immune variations in bipolar disorder: phasic differences. Bipolar. Disord. 9:596–602.
Wadee AA, et al. (2002) Serological observations in patients suffering from acute manic episodes. Hum. Psychopharmacol. 17:175–9.
Lucas SM, Rothwell NJ, Gibson RM. (2006) The role of inflammation in CNS injury and disease. Br. J. Pharmacol. 147 Suppl 1:S232–40.
Myint, et al. (2007) Effect of the COX-2 inhibitor celecoxib on behavioural and immune changes in an olfactory bulbectomised rat model of depression. Neuroimmunomodulation 2007;14:65–71.
Dausmann KH, Glos J, Ganzhorn JU, Heldmaier G. (2004) Physiology: Hibernation in a tropical primate. Nature 429:825–6.
Prendergast BJ, Freeman DA, Zucker I, Nelson RJ. (2002) Periodic arousal from hibernation is necessary for initiation of immune responses in ground squirrels. Am. J. Physiol. Regul. Integr. Comp. Physiol. 282:R1054–62.
Muller N, et al. (2006) The cyclooxygenase-2 inhibitor celecoxib has therapeutic effects in major depression: results of a double-blind, randomized, placebo controlled, add-on pilot study to reboxetine. Mol. Psychiatry 11:680–4.
Nery FG, et al. (2008) Celecoxib as an adjunct in the treatment of depressive or mixed episodes of bipolar disorder: a double-blind, randomized, placebo-controlled study. Hum. Psychopharmacol. 23:87–94.
Acknowledgments
The study was supported by the Max Planck Society, by several private donations, as well as by the DFG Center for Molecular Physiology of the Brain (CMPB). The authors would like to thank Jeannine Dietrich and Kathrin Hannke for excellent assistance and Fred Wolf for helpful discussions.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
10020_2008_1409546_MOESM1_ESM.pdf
Episode-Specific Differential Gene Expression of Peripheral Blood Mononuclear Cells in Rapid Cycling Supports Novel Treatment Approaches
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Begemann, M., Sargin, D., Rossner, M.J. et al. Episode-Specific Differential Gene Expression of Peripheral Blood Mononuclear Cells in Rapid Cycling Supports Novel Treatment Approaches. Mol Med 14, 546–552 (2008). https://doi.org/10.2119/2008-00053.Begemann
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.2119/2008-00053.Begemann