Abstract
Background
Taichung Native 1 (TN1) is the first semidwarf rice cultivar that initiated the Green Revolution. As TN1 is a direct descendant of the Dee-geo-woo-gen cultivar, the source of the sd1 semidwarf gene, the sd1 gene can be defined through TN1. Also, TN1 is susceptible to the blast disease and is described as being drought-tolerant. However, genes related to these characteristics of TN1 are unknown. Our aim was to identify and characterize TN1 genes related to these traits.
Results
Aligning the sd1 of TN1 to Nipponbare sd1, we found a 382-bp deletion including a frameshift mutation. Sanger sequencing validated this deleted region in sd1, and we proposed a model of the sd1 gene that corrects errors in the literature. We also predicted the blast disease resistant (R) genes of TN1. Orthologues of the R genes in Tetep, a well-known resistant cultivar that is commonly used as a donor for breeding new blast resistant cultivars, were then sought in TN1, and if they were present, we looked for mutations. The absence of Pi54, a well-known R gene, in TN1 partially explains why TN1 is more susceptible to blast than Tetep. We also scanned the TN1 genome using the PosiGene software and identified 11 genes deemed to have undergone positive selection. Some of them are associated with drought-resistance and stress response.
Conclusions
We have redefined the deletion of the sd1 gene in TN1, a direct descendant of the Dee-geo-woo-gen cultivar, and have corrected some literature errors. Moreover, we have identified blast resistant genes and positively selected genes, including genes that characterize TN1’s blast susceptibility and abiotic stress response. These new findings increase the potential of using TN1 to breed new rice cultivars.
Similar content being viewed by others
Background
The Green Revolution (GR) in rice production was attributed to the high-yielding semi-dwarf cultivars. In fact, the miracle rice, IR8, inherited the sd1 (semidwarf 1) gene from the Dee-geo-woo-gen (DGWG) cultivar (Hargrove et al. 1979). It conferred IR8 its short stature, making it lodging resistant, leading to high grain yield. Unknown to many, another cultivar also inherited the sd1 gene directly from DGWG. It is the Taichung Native 1 (TN1), which was popular in the 1960s (Chandler 1992). Recently, the genome of TN1 was sequenced, assembled and annotated, hel** to answer questions about the yield difference between TN1 and IR8 and why they both are photoperiod-insensitive (Panibe et al. 2021).
A fundamental characteristic of TN1 is its short height due to the sd1 gene from DGWG. The deletion of the semidwarf sd1 gene incurs a loss of function for the gibberellin (GA) 20-oxidase 2 (Os20ox2), which is involved in the synthesis of the growth hormone gibberellin (Spielmeyer et al. 2002). A reduction in GA results in a shorter plant height (Itoh et al. 2002). However, the sequence of the sd1 gene is not well studied. The current literature definition of the sd1 gene was based on the comparison of DGWG-type sd1 mutants (Habataki, Milyang 23, and IR24) with the sd1 of Nipponbare, Sasanishiki, and Calrose (Monna et al. 2002). It revealed a 383-bp deletion from the second half of Nipponbare’s exon 1 to the first half of exon 2, or in terms of the expressed sequence, a 278-bp deletion (Monna et al. 2002). Another definition of the sd1 deletion is a 280-bp deletion in the comparison of the semidwarf Doongara with the tall Kyeema, whose sd1 sequence is similar to Nipponbare (Spielmeyer et al. 2002). Those studies were done when the full Nipponbare genome was not yet available (until 2005) (International Rice Genome Sequencing Project and Sasaki 2005), and was later improved in 2013 (Kawahara et al. 2013). With the genomes of TN1 (Panibe et al. 2021) and IR8 (Stein et al. 2018) now available, we aim to compare the sd1 genes of these cultivars and redefine the semidwarf gene based on TN1 and IR8, the two direct descendants of DGWG.
If the greatest strength of TN1 is its high-yielding property due to its semi-dwarf stature from the sd1 gene, its weakness is its high susceptibility to the blast disease. Rice blast leads to a severe annual loss in rice production worldwide (Wang et al. 2014). However, plants have a natural defense against this and other pathogens, thanks to their resistance genes or R genes. Most R genes are composed of a nucleotide-binding site (NBS) domain and a leucine-rich repeat (LRR) domain (Takken and Joosten 2000). A combination of R genes in a plant may lead to a wide range of immunity response (Fukuoka et al. 2015). Unfortunately, TN1 is susceptible to major rice diseases like blast caused by the fungus Pyricularia oryzae (syn. Magnaporthe oryzae) (Sabbu et al. 2016) and the bacterial blight disease caused by the bacteria Xanthomonas oryzae pv. oryzae (Kumar et al. 2012). Predicting the R genes in the genome of TN1 will help understand the resistance profile of TN1, and why it is highly susceptible to blast. For factors that affect plant sensitivity to blast disease, see Chen et al. (2019), Liu et al. (2021), Nugroho et al. (2021) and Zhang et al. (2015).
There are in total 37,526 predicted genes in the TN1 genome (Panibe et al. 2021). Of these thousands of genes, some could be under the influence of positive selection (PS), conferring the cultivar certain advantages that could be related to TN1’s phenotypic characteristics like drought tolerance (Garg and Singh 1971; Garg et al. 2002). Mining the entire genome for genes that makes TN1 unique is no longer highly challenging, thanks to bioinformatics tools that automate the process of looking for positively selected (PS) genes such as PosiGene (Sahm et al. 2017). By using an input of coding sequences from the genomes of GR-related cultivars like IR8 (Stein et al. 2018), MH63 (Zhang et al. 2013) by using the protein alignments of sd1 and the information from their gff annotation (Nagano et al. 2005; Panibe et al. 2021). The range specified by the light blue arrow represents the sequences of sd1 in TN1 and IR8 that were validated by our Sanger sequencing. The 382 bp deletion in TN1 can be derived by computing the difference between 981 and 599, the latter of which represents the gene length of TN1 sd1 before its 2nd intron
We further confirmed the sd1 gene sequence of TN by map** TN1 short reads used in the 3000 Rice Genomes Project (3 K RGP) (Wang et al. 2018b). There are actually two sets of TN1 reads in the 3000 Rice Genomes Project and they have the assay IDs, CX270 and CX162. The former has the name TAICHUNGNATIVE1, while the latter is designated as TN1. To determine which one better represents the sequencing reads from the 3000 Rice Genomes Project, we mapped the reads to the TN1 genome. CX162 has a 99.92% and 90.92%, for the overall map** rate and properly paired mapped reads, respectively. In contrast, CX270 has a map** rate of 99.40% and 81.91%. Based on the map** of reads, CX162 better represents the TN1 genome in the 3 K RGP.
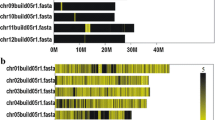
We also checked the SNP-Seek database (Mansueto et al. 2017), if there are SNP loci inside the region corresponding to the sd1 deleted sequence in semi-dwarf cultivars. Of the two, TN1 (CX162) has missing SNP positions to deletion in japonica (Fig. 3a), whereas TAICHUNGNATIVE1 (CX270) has alleles on the same set of coordinates. (Additional file 1: Fig. S3). We further inspected the map** of the reads by viewing the sd1 region in Integrative Genomics Viewer (IGV) (Robinson et al. 2011), and they are shown in Fig. 3b (CX162) and Additional file 1: Fig. S4 (CX270). The nucleotide at chromosome 1 position 40,362,230 was supported by the TN1 reads of CX162 (Fig. 3c) and CX270 (Additional file 1: Fig. S5). The former’s reads better covered the position compared to the latter. In CX162, it is mapped by six reads, while in CX270 it is by only one read.
sd1 gene SNP-Seek result for TN1 and map** of the CX162 reads. a Screenshot of the Genotype search result of the SNP-Seek database for the sd1 gene. The blank green space for the TN1 cultivar assay CX162 signifies the absence of genomic DNA in TN1 with respect to the Nipponbare reference genome. According to the alignment in Fig. 1, the deleted region in Nipponbare sd1 lies in between 38,382,762 and 38,383,144 of chromosome 1 and is consistent with the missing TN1 SNPs in SNP-Seek with respect to the Nipponbare reference genome starting at position 38,382,846 up to position 38,383,066. b Map** coverage of the TN1 (assay CX162) 3 K RGP reads onto the sd1 gene region of the TN1 genome. The bam file was filtered to only get the properly paired reads with the proper distance. The gray color means that the read sequence is identical to the TN1 genome. The red asterisk is the approximate location of the start of the sd1 deletion with respect to the Nipponbare genome. c Closeup of the asterisk area in Fig. 4b. It has the coordinate 40,362,230 in TN1 chromosome 1. It corresponds to the extra nucleotide that made the sd1 deletion in TN1 and IR8 382-bp, instead of 383-bp. It is covered by 6 properly mapped paired-end reads with the correct distance. Gray color means the read sequence are homozygous to the TN1 genome
Predicted R genes in TN1
We annotated 383 NLR (nucleotide-binding domain leucine-rich repeat), 34 NB-ARC and 6 LRR (leucine-rich repeat) in the TN1 genome (Additional file 2: Dataset S1). For this purpose, we used the Tetep as a reference because Tetep is known to be highly resistant to blast disease and its genome and R genes have been well characterized (Wang et al. 2019b); indeed, it has been commonly used to breed for new blast resistant cultivars (Singh et al. 2012; Zarbafi and Ham, 2019; Ramalingam et al. 2020). The numbers of orthologues found between Tetep (Wang et al. 2019b) and TN1, MH63, R498 and Nipponbare did not show significant differences (Additional file 1: Table S1). Non-orthologous Tetep NLRs (R genes) were then blasted against the TN1 proteome using their NR-ARC domain protein sequences (Additional file 3: Dataset S2) and those hits with alignment identity < 50% were deemed missing in the TN1 assembly. One of the unfound R genes in TN1 is Pi54 (Pik-h), which is the gene chr11.fgenesh2107 in the assembled Tetep genome (Wang et al. 2019b). Pi54, originally cloned from Tetep, is known to confer broad-spectrum resistance to blast (Gupta et al. 2011; Rai et al. 2011; Thakur et al. 2015). Moreover, ~ 28 of the 90 NLR genes that were found to be resistant to one or more blast fungal strains (Wang et al. 2019b) were found to be missing or mutated in the TN1 genome (Additional file 3: Dataset S2).
By using the method of Mahesh et al. (2016), the set of 22 cloned blast R genes were searched in the TN1 genome. The results are given in Table 1 and those marked with an asterisk were the results different from Mahesh et al. (2016). These genes are confirmed to be present by Blastp in the Tetep genome with the same criteria used by Mahesh et al. (2016), i.e., e-value < 10e-10, identity ≥ 70% and query coverage ≥ 70% (Additional file 4: Dataset S3). The same set of R genes were also searched in the TN1 genome. Some of the R genes are present in TN1 but are mutated (Table 1), preventing the translation of the gene into the right protein. In the case of Tetep, both Wang et al. (2019b) and Mahesh et al. (2016) found the Pi-ta and Pi54 R genes in the blast resistant cultivar (see the Tetep column in Table 1).
Table 1. Distribution of cloned blast resistance genes in sequenced rice varieties. A + means present and a− means absent while M means mutated but with protein structure retained. *means a result different from Mahesh et al. (2016). These genes are confirmed to be present by blastp in the Tetep (Wang et al. 2019b) genome with e-value < 10e−10 and identity > = 70% (see Additional file 4: Dataset S3 for details). chr11.fgenesh2107.1 is a gene name from the Tetep genome annotation.
Haplotype analysis of the Pi-ta and Pi54 genes
To filter the missense variants, we compared each allele in the haplotypes of Pi54 and only obtained the SNPs that are heterozygous (see “Methods” section). Only three SNP positions were left and they are located on chromosome 11: 25,263,636; 25,264,119; and 25,264,164 (see Table 2). We checked their allele frequencies and found that the missense variants have a minimum allele frequency (MAF) of 36% to 39% (Table 2), suggesting that these missense variants are maintained across the rice populations, even though each causes a change in amino acid. Using the major/minor allele section in Table 2, we compared the three alleles of TN1 to the major and minor alleles. At SNP position 25,263,636, TN1 has two possibilities: either allele C or T (Table 2). If it is a T, it will be a minor allele across the 3,024 rice cultivars. The missense variant causes a Glu144Lys mutation (Additional file 6: Dataset S5), changing an acidic amino acid into a basic one. The change in charge of an amino acid could disrupt ionic interactions in the structure of the protein, which could affect its function, supporting our observation from Table 1 that the Pi54 gene in TN1 is missing when compared to the blast resistant Tetep cultivar.
We also investigated the Pi-ta gene in SNP-Seek and it returned four haplotypes (Additional file 5: Dataset S4). This is the same number of haplotypes that Jia et al (2003) found. In that study, three of the haplotypes were related to susceptibility to blast and have five nucleotide positions that caused a non-synonymous mutation in Pi-ta. We checked the annotation of the SNPs in the 3 K RGP and found that the I6S mutation (Additional file 6: Dataset S5) was due to the replacement of the G nucleotide by a T at position 10,611,754 (Table 3). TN1 has the A allele at this position, which is the minor allele across the 3,024 cultivars in SNP-Seek. Consequently, TN1 is predicted to have the I6S mutation in its Pi-ta protein. From Table 3, the resistant cultivars Katy and Drew have the alleles T, G and C at positions 10,611,244; 10,611,297; 10,611,327, and an A at position 10,611,754.
However, the susceptible Nan**g 11 cultivar as well as TN1 has the pattern of alleles at the mentioned SNP positions similar to Katy and Drew. We did not see very clear difference between the haplotypes of the susceptible ones and the resistant ones for Pi-ta (Table 3) but for Pi54 the differences look like a bit clearer (Table 2). Pi54 is considered non-functional as the allele in TN1 (OsTN11t002257.1) lost the first 598 amino acids when compared to Tetep (Additional file 1: Fig. S6), resulted in complete loss of the NB-ARC domain. Of the 9 absent cloned NLRs (11 alleles shown in Table 1, which belong to 9 genes) in TN1, only 3 are from indica donors (other 6 might represent japonica/indica differences), including Pi54, Pid2 and Pi1-5. Pid2 is present in both susceptible and resistant cultivars, while Pid1-5 is absent in them all. Only Pi54 shows presence/absence polymorphism in resistant (Tetep and Tadukan) and susceptible (Co-39 and HR-12) cultivars (Mahesh et al. 2016). We further investigated the Pi-ta gene of TN1 by aligning it against its counterpart in Yashiro-mochi (a resistant cultivar). The protein sequence alignment of Pi-ta in TN1 is largely the same compared to the latter (Additional file 1: Fig. S7), suggesting that the function of the gene in TN1 is not largely altered.
Eleven genes in TN1 underwent positive selection
The aim of the genome-wide search for TN1 genes that underwent positive selection (PS) is to identify genes that might explain TN1’s phenotypic characteristics like high yield (Yoshida 1981), photoperiod insensitivity (Vergara and Chang 1985) and drought-tolerance (Garg and Singh 1971). The GO terms assigned to these PS genes would give insights into the biological processes involved as well as the enzymes that confer the function. We identified 11 TN1 genes that were likely subject to PS in TN1 in the past (Table 4).
Using the Blast2GO annotation of the TN1 assembly (Panibe et al. 2021), a total of 35 GO terms (Additional file 1: Table S3) were assigned to six of the 11 PS genes (Table 4); see their representative GO terms in Fig. 4. For the Molecular Function (Fig. 4b), a correlation is observed between the protein names of the six genes and their GOs.
REVIGO (Supek et al. 2011) visualization of GOs of genes under positive selection in TN1. a Biological process and b molecular function. These are the scatterplot of REVIGO showing the representative GO terms of the PS genes, where the colors of the circles represent the uniqueness value, computed from comparing each GO term to each other. The bigger the size of the sphere, the more the general the term is